Unveiling the release mechanism of small extracellular vesicles with an unprecedented throughput
Published in Bioengineering & Biotechnology, Protocols & Methods, and Cell & Molecular Biology

Small extracellular vesicles (sEVs) mediate intercellular communication by delivering bioactive molecules in our body and are increasingly attracting attention as naturally derived drug delivery vehicles. Aiming at developing “designer” sEVs for efficient cargo delivery by engineered sEVs, we previously developed synthetic-biology-inspired “EXOtic devices” for the efficient production of sEVs loaded with RNA of interest [1]. Through this study, while we were convinced that the sEVs are promising as therapeutic modalities, we also felt that a deep understanding of sEV biology is necessary for further exploiting the potential of sEVs for future applications. This feeling was the driving force for us to establish a new exploratory system to unveil the genes/proteins controlling the release of the sEVs.
To deepen our understanding of sEVs, we wanted to develop a high throughput system to unveil the controller of sEVs without prior knowledge. When we started our project, the pooled CRISPR screening system, which allows researchers to perform genome-wide screening of regulators of various biological phenomena depending on the experimental setup, had been increasingly becoming popular. We thought it would be great to incorporate this system in our study, though our initial question was how to apply this to sEV research. In normal CRISPR screening, the gRNA-encoding region transduced in the cellular genome is used for high throughput counting of the cells, each in which a different gene is knocked out. We hypothesized that transcribed gRNA could also be used for this purpose and came up with combining this idea and our EXOtic devices to encapsulate specific RNA into sEVs; by encapsulating transcribed gRNA into sEVs as “barcodes” in a format of pooled CRISPR screening, we anticipated that the amount of sEVs released from each cell can be estimated by analyzing the barcode composition in the sEVs by next-generation sequencing (NGS) (Figure 1).
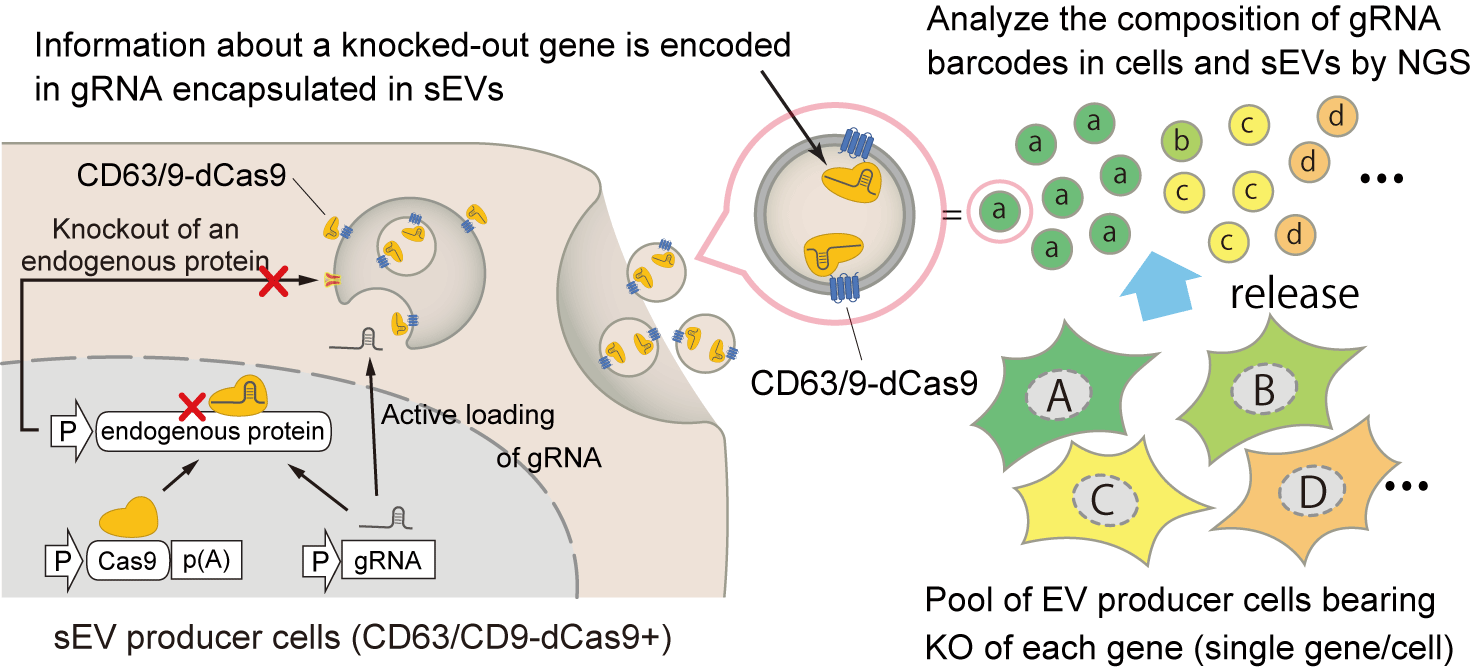
Figure 1: Concept of the CIBER screening system. CRISPR gRNA is actively encapsulated into the sEVs by the interaction with dCas9 fused with an sEV marker. By analyzing the barcodes in the sEVs and cells by NGS, release regulators of sEVs can be analyzed in a pooled manner.
Our initial strategy was recruiting gRNA bearing a MS2 loop, an RNA motif that binds to its partner protein MS2 coat protein (MCP), into sEVs by using its interaction with MCP fused with a sEV marker CD63 (CD63-MCP) because gRNAs bearing MS2 loops had been already used for CRISPR activation screening [2]. The first hurdle was to develop a protocol to amplify gRNA for NGS readout. At the time, no researcher had tried sequencing transcribed gRNA, and we found that this is not easy without optimization because the spacer sequence of the Cas9 gRNA is located at the 5’ end and normal PCR cannot be applied. We found that the incorporation of SMART-Seq2 technology [3] would be a good solution for this, and we continued optimizing the protocol. However, even after making significant improvements to the protocol, we realized that the barcoding efficiency by CD63-MCP and MS2-gRNA never reached a sufficient level (40% of gRNA was found to be missing) in our settings, and also the distribution of barcode coverage in sEVs was highly skewed. The breakthrough here was that we came up with the idea to use dead Cas9 (dCas9) as the gRNA-recruiting protein instead of MCP. With this system, we succeeded in routinely getting rich coverage of gRNA (approximately 99%) displaying low skewness. It was gratifying that the necessary cell culture scale for getting such barcoding efficacy was not much different from that of normal CRISPR screening. Another merit of this system is that we can use a normal gRNA scaffold, allowing for the use of commercially available gRNA libraries without modification.
While the analytic pipeline of normal pooled CRISPR screening is well established after getting the raw read count of each gRNA, we struggled with establishing the analysis process; it was not preferable to determine the gene rank by simply comparing the read count of gRNA in sEV between control and knockout conditions because we found that the amount of gRNA in the sEV fraction was significantly affected by that in the cellular fraction. It was fortunate that we could collaborate to overcome this issue with Dr. Tadahaya Mizuno, who is an expert in bioinformatics. After much discussion, we managed to devise the calculation method that can reliably evaluate the changes in sEV release upon gene knockout while canceling out the effect of cellular activity (viability, barcode transcription, etc., see Fig. 2 in the original paper), which became an indispensable analytical pipeline throughout the study. (As a side note, Kojima was personally very happy that the collaboration was done with Dr. Mizuno because they have been old friends since their university days…!)
After naming the screening pipeline as “CIBER screening,” we were dedicated to downstream analysis, but this was also not that simple. While it is great that CIBER screening provides massive information on potential sEV release regulators, it was rather difficult to extract biologically important information from screening results. This was partly because we are tool developers in a chemistry-oriented lab and quite unfamiliar with the deep depths of pure biology. There were two keys; what biological phenomena to focus on and how to analyze the substantial data. For the former, while we had too many possible research directions and had few clues to decide where to go at the beginning (it was like being trapped in the deep fog of sEV biology…), discussions with the active researchers in sEV field in Japan through the JST PRESTO/CREST program as well as reading a couple of pioneering works showing the importance of taking the sEV subpopulation into consideration (such as a reference [4]) were quite helpful to determine the direction of our research. For the latter, we appreciated the power of bioinformatic analysis for exploratory purposes. The remaining question was how to extract biologically important information from the screening data, each of which inevitably contains some degree of noise. Here, Dr. Mizuno suggested that the integrated analysis of all the data of individual genes without arbitrariness will provide us with higher-order, bird’s-eye views with important insights. Indeed, Gene Ontology analysis and a Gene Set Enrichment Analysis (GSEA) beautifully depicted important biological pathways even from screening the score of individual genes not very clear at a glance. Of course, more importantly, we are happy that we could experimentally prove this finding, revealing the previously unknown biological processes that had differently affected different subpopulations of sEVs. (Please read the paper for the details!)
One of the irreplaceable contributions of experimental scientists in the era of AI is to obtain valuable primary data bearing rich contents. From this viewpoint, we believe CIBER matches with the time, providing massive information on sEV biology in an unprecedented throughput, which can be analyzed from various aspects as one’s capability of data processing advances in the future. Conducting CIBER screening under various conditions with multiple cell lines would be an effective approach for unveiling other previously unknown factors controlling sEV release, so we hope multiple researchers try the CIBER screening system with their hands and provide useful information from different aspects. In this sense, the direct compatibility of commercially available gRNA to the CIBER system helps researchers perform screening without big barriers. Also, as discussed in the paper, barcoded sEV could be used to estimate cell population dynamics without destroying cells, and tracing the fate of barcoded sEVs in various contexts would help us better understand sEV biology. Thus, marking the current work as an important milestone, we are excited to set off on our next adventure with the system!
References:
1.Kojima, R. et al. Designer exosomes produced by implanted cells intracerebrally deliver therapeutic cargo for Parkinson’s disease treatment. Nat. Commun. 9, 1305 (2018).
2. Konermann, S. et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517, 583–588 (2015).
3. Picelli, S. et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods 10, 1096–1098 (2013).
4. Mathieu, M. et al. Specificities of exosome versus small ectosome secretion revealed by live intracellular tracking of CD63 and CD9. Nat. Commun. 12, 4389 (2021).
Our paper:
Koki Kunitake, Tadahaya Mizuno, Kazuki Hattori, Chitose Oneyama, Mako Kamiya, Sadao Ota, Yasuteru Urano, Ryosuke Kojima*
Barcoding of small extracellular vesicles with CRISPR-gRNA enables comprehensive, subpopulation-specific analysis of their biogenesis and release regulators
Nature Communications 2024, 15, 9777.
URL: https://www.nature.com/articles/s41467-024-53736-x
Acknowledgements and credit for the banner image
We thank Dr. Tadahaya Mizuno and Ms. Winnie Wong for useful comments on this manuscript. Credit of the graphic should be addressed to Koki Kunitake.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in