Using clusterProfiler to characterise Multi-Omics Data
Published in Cell & Molecular Biology, Genetics & Genomics, and Statistics

ClusterProfiler was developed over a decade ago and first published in OMICS in 2012. It was created to address the emerging need for complex experiment designs involving multiple time points and conditions, as simple case-control setups were predominant at that time. The package has continuously evolved, featuring an exceptionally user-friendly interface. It has now become a popular tool in the bioinformatics community.
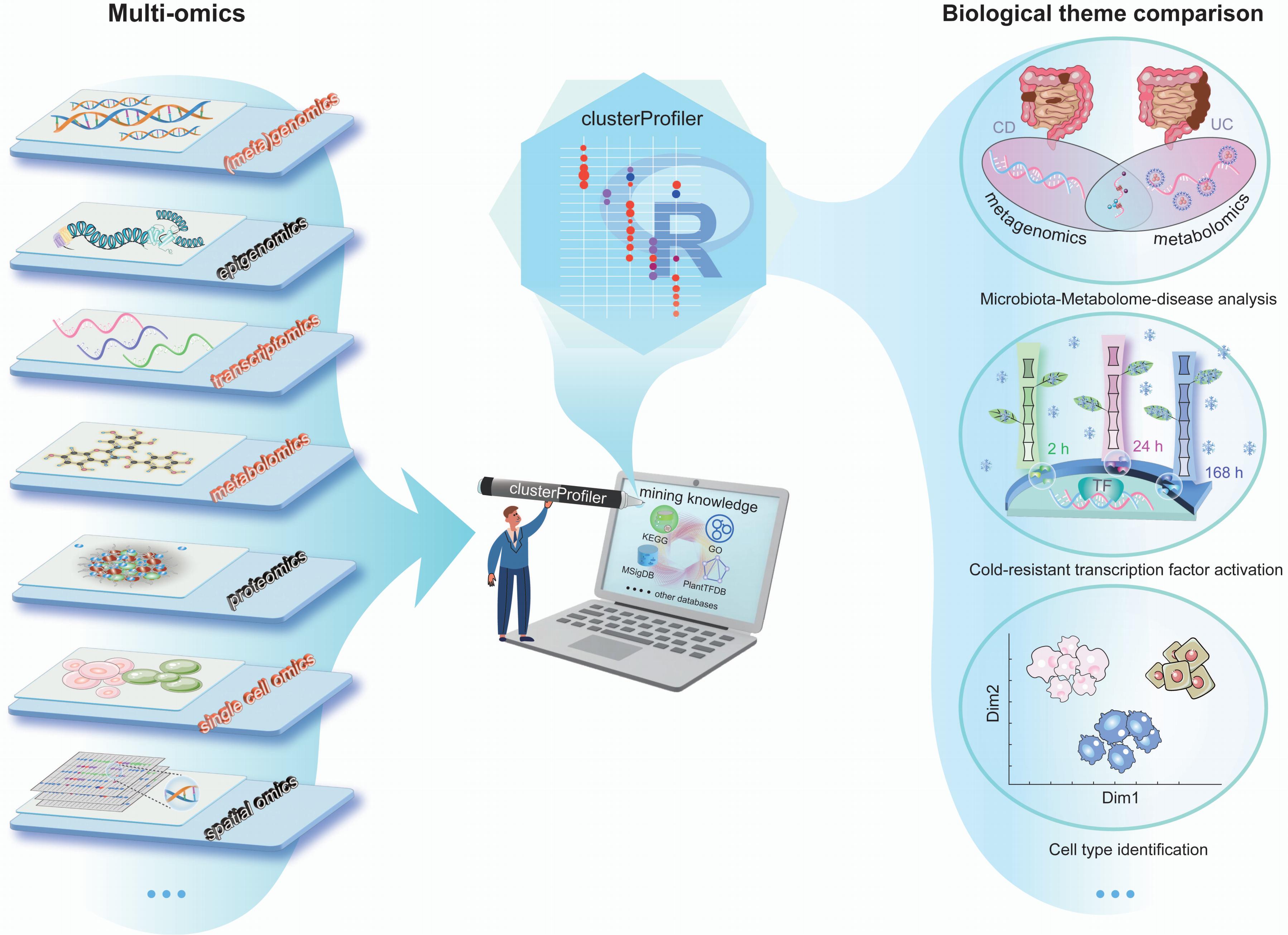
ClusterProfiler offers several key features that make it stand out from similar tools. It facilitates online retrieval of the latest genome annotation, supporting Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis for thousands of species. Additionally, it provides a universal interface for incorporating user-provided custom annotation, enabling analysis of new species and utilization of new functional annotation. Notably, clusterProfiler allows for the utilization of genomic regions, enabling enrichment analysis of epigenomic data. Moreover, it implements comparative analysis of multiple datasets to support complex experimental designs and offers a tidy interface, streamlining data manipulation and interpretation.
This protocol mainly introduces three user cases, demonstrating the functionality of comparing biological themes in disease subtypes, analyzing functions of perturbed transcription factors, and annotating cell types using various omics data sets, including metagenomics, metabolomics, transcriptomics, and single-cell omics.
It is not limited to the types of omics data presented and can be used to uncover potential molecular mechanisms from the user's chosen perspective.
References:
- S Xu#, E Hu#, Y Cai#, Z Xie#, X Luo#, L Zhan, W Tang, Q Wang, B Liu, R Wang, W Xie, T Wu, L Xie, G Yu*. Using clusterProfiler to characterise Multi-Omics Data. Nature Protocols. 2024, accepted.
- T Wu#, E Hu#, S Xu, M Chen, P Guo, Z Dai, T Feng, L Zhou, W Tang, L Zhan, X Fu, S Liu, X Bo*, G Yu*. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. The Innovation. 2021, 2(3):100141.
- G Yu, LG Wang, Y Han, QY He*. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS: A Journal of Integrative Biology. 2012, 16(5):284-287.
- Exploring Biological Knowledge and Discovery · YuLab@SMU (yulab-smu.top)
Follow the Topic
-
Nature Protocols

This journal publishes secondary research articles and covers new techniques and technologies, as well as established methods, used in all fields of the biological, chemical and clinical sciences.



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in