Validation of MSIntuit as an AI-based pre-screening tool for MSI detection from colorectal cancer histology slides
Published in Cancer

With the advent of precision medicine, characterising the genotype of a cancer tumor is becoming more important to determine how oncologists treat it. This is the case in colorectal cancer, where cancer cells’ DNA mismatch repair (MMR) systems can become faulty. This leads to errors, such as insertions and deletions, appearing in the cells’ DNA. This genomic condition is known as Microsatellite Instability (MSI).
Thankfully, MSI makes tumor cells more susceptible to immunotherapy. There is now even immunotherapy treatment approved specifically for patients whose cancer tumors display MSI; for example pembrolizumab, a PD-1/PD-L1 inhibitor that targets immune cells. So MSI testing is now a crucial part of colorectal cancer treatment decision making.
There are two main ways to test MSI: immunohistochemistry (IHC), to detect loss of MMR proteins, or molecular tests such as polymerase chain reaction (PCR) to show microsatellite alterations. Both of these methods have drawbacks:
|
IHC |
PCR |
|
Requires excellent tissue fixation |
Requires specific PCR machines that not all centres have |
|
Slide preparation time |
Long turnaround time |
|
Consumes already scarce tissue material |
Laboratory and technicians required |
|
Requires experienced pathologist of which there is currently a global shortage |
This is the backdrop to Owkin’s work on MSIntuit™ CRC, an AI diagnostic that pre-screens for MSI in colorectal cancer patients using only routinely collected H&E slides that have been digitized. The tool rules out 40% of colorectal cancer patients from needing IHC or PCR testing for MSI, which saves both lab testing resources and pathologists’ time. This is the first clinically approved AI-based tool for MSI detection from H&E slides. 
An overview of MSIntuit™ CRC use in clinical practice
We started research on this project in 2018. We wanted to design an algorithm that could recognize MSI-status from H&E images. At this point, the project was more of a scientific proof-of-concept: could an algorithm recognize this tissue’s genotype from looking at the histology alone? Our initial models worked well, but the real breakthrough came in 2019. This is when we began incorporating self-supervised learning (SSL) techniques into our models, to discriminate specific visual features in the image using a vast amount of unlabeled histology images. Those features could then be associated with genotype. This brought our algorithm to high enough levels of accuracy for us to consider turning it into a diagnostic product.
| Pre-training dataset | Method | AUROC on PAIP | AUROC on MPATH-DP200 |
| ImageNet | Supervised | 0.92 [0.84- 0.97] | 0.79 [0.74-0.83] |
| TCGA | SSL (MSIntuit) | 0.96 [0.90- 0.99] | 0.88 [0.84-0.91] |
MSIntuit™ CRC approach, which relies on SSL, substantially outperforms a common approach relying on ImageNet supervised learning on two external datasets
But to work as a tool for clinical practice, we needed to ensure three key things. First, that our model was as sensitive (or more) to MSI as existing testing techniques. Second, that our model had a high enough degree of specificity that it could rule out a useful number of non-MSI (MSS) patients. Third, that the model could generalize across pathology labs equipped with different scanners. This is the purpose of our recently published Nature Communications paper.
To address these points, we performed a blind clinical validation of MSIntuit™ CRC on a cohort of 600 colorectal cancer cases. One of the major challenges was to make sure that for each pathology lab where the tool is deployed, a high sensitivity is obtained, even under the presence of domain shift. Most studies of AI tools focus on the area under the ROC curve (AUROC) as their main performance metric, neglecting this important point. In our study, we used an innovative calibration step to ensure the tool’s sensitivity holds across pathology labs. It consists of using a small number of MSI slides from the lab where the tool will be deployed to determine the operating threshold that yields a high sensitivity.
The final results showed a sensitivity of 97% and 95%, and a specificity of 46% and 47% across the same set of histology images, digitized with two different scanners - a sensitivity in line with standard screening methods.
The model’s results on both scanners were statistically very similar, highlighting its robustness to the scanner used. Then we tested its intra-scanner specificity by digitizing 30 slides 8 times on the same scanner. Again the model’s results on all sets of data were statistically comparable. This was a hugely satisfying result as it suggests MSIntuit™ CRC is ready to be used in clinical practice - and in fact it is now being rolled out over a number of pathology laboratories in France.
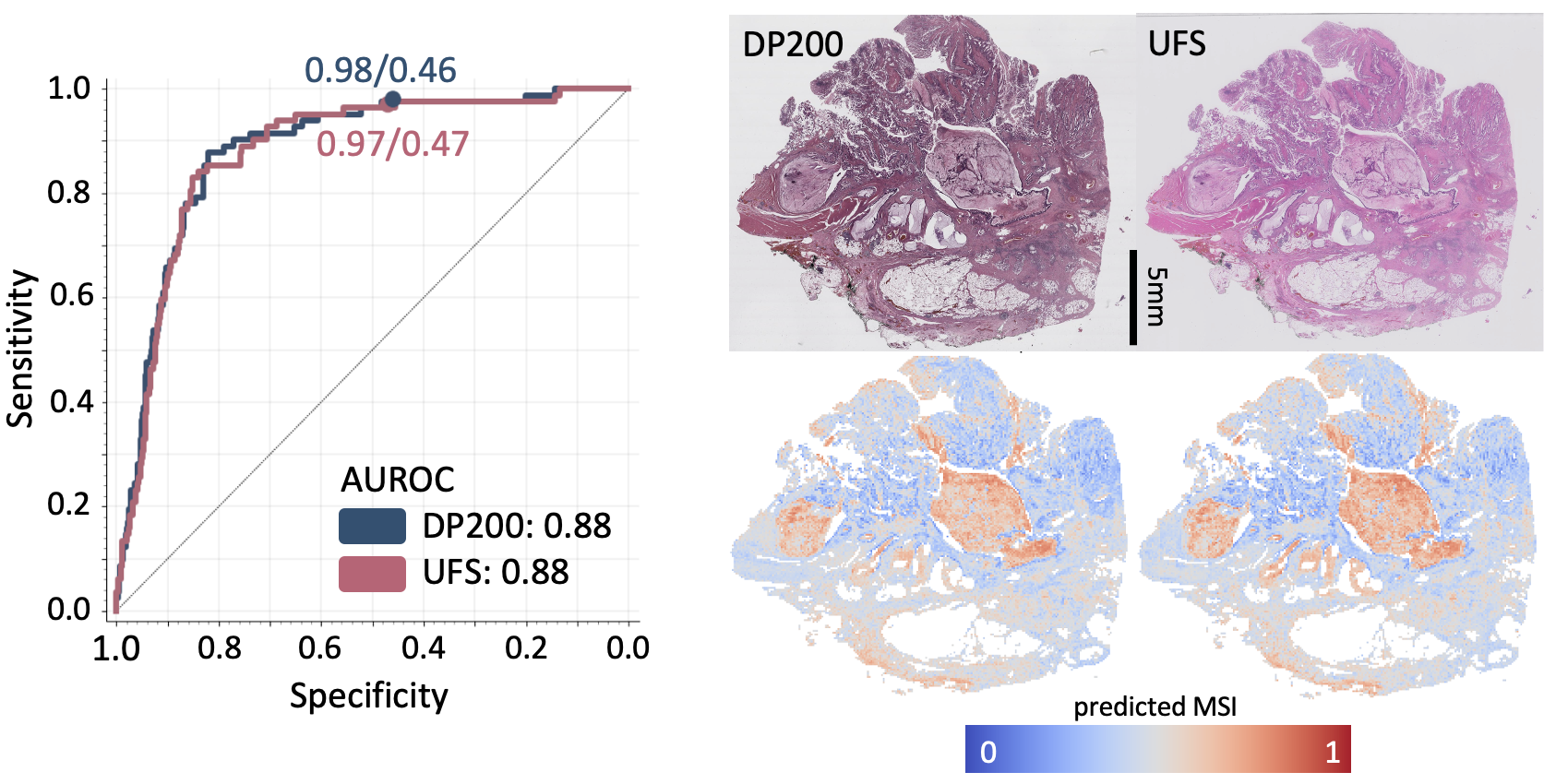 Left: Performance of MSIntuit™ CRC (ROC curves) on the two different scanners. Right: An H&E slide digitized on the two different scanners and the corresponding MSIntuit™ CRC heatmaps.
Left: Performance of MSIntuit™ CRC (ROC curves) on the two different scanners. Right: An H&E slide digitized on the two different scanners and the corresponding MSIntuit™ CRC heatmaps.
Cancer treatment, and indeed medicine more broadly, is moving towards more precisely targeted disease treatment. This kind of precision medicine aims to target specific subgroups of patients with the treatment they will most benefit from. As we move forward, diagnosing patient subgroups will become increasingly important in determining treatment. This is where we believe that such AI-based pre-screening tools will be vital.
The ability to predict the presence of biomarkers like MSI, that correspond to patient subgroups, from H&E slide images alone, is one of the standout applications of AI. If these tools can be designed to be reliable and generalizable (as we have found here), we believe they can make diagnosis of high volumes of biomarkers sustainable, to support precision medicine. We also hope that the relative ease and cost-effectiveness of the solution can help to democratize access to this kind of diagnosis in low resource settings.
Conflicts of interest: I am an employee of Owkin, and MSIntuit™ CRC is a tool commercialized by Owkin.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in