Viral-bacterial co-infections screen in vitro reveals molecular processes affecting pathogen proliferation and host cell viability
Published in Protocols & Methods, Biomedical Research, and Immunology
Infectious diseases pose a severe threat to public health and have recently moved back into the focus of both the scientific research community, as well as the broad public. This is most strongly caused through the SARS-CoV-2 pandemic, stifling public life across the globe and causing additional rippling effects. Beyond that, the emergence of antibiotic-resistant bacterial pathogens, compounded by the slow-down of the development of novel antibacterial treatments, and the continuous expansion of the endemic areas of tropical diseases create the need to better understand how pathogens affect us and how we interact with them.
When our body encounters a pathogen, our cells initially respond though the initiation of innate immune defenses. These comprise signaling cascades that alert neighboring cells and activate the immune system to combat the infection, for example through the recruitment of cells that can kill pathogens, such as macrophages. Through thousands of years of co-evolution, pathogens have developed techniques to hijack the innate immune response and turn it to their advantage, making this a crucial interaction point between us – the host – and the pathogen. The host-pathogen interface is even more intricate in the case of two or more co-occurring pathogens. There are abundant reports about co-infections exacerbating symptoms for patients, causing increased mortality, especially in children, the elderly and the immunocompromised. Yet on a cellular or molecular level we know very little about how pathogens interact with each other and how our cells mediate the processes that cause this increased disease severity.
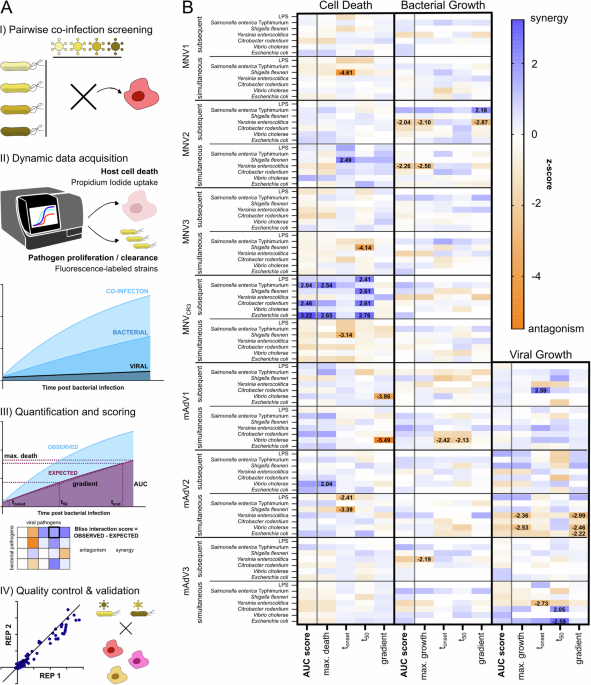
We therefore wanted to explore the mechanisms that are at the basis of the interface between the host and the two pathogens. In this study, we provide a first systematic assessment of interactions between enteric viral and bacterial pathogens. To achieve this, we developed a simplified model system that allows us to assess a large number of pathogen pairs in parallel and over time. We quantify host cell death and pathogen growth or clearance as initial metrics. After ensuring the validity of this large dataset, as well as additional quality control, we dive deeper into the molecular mechanisms and uncover two very different interaction points for the viral and bacterial invader.
Many infectious agents need to invade the host cell to propagate and spread. The first interaction point we focus on was therefore on cell entry: Does one pathogen make it more or less likely that a second pathogen gets eaten by macrophages? We discover that an acute strain of murine Adenovirus (mAdV2) causes mouse macrophages to more efficiently take up Yersinia, a bacterium that causes enteritis and colitis, while other bacterial pathogens were unaffected. This is interesting since Yersinia actively prevents being taken up and destroyed by macrophages. We could identify that the viral infection causes an increased production of proteins that rearrange the cytoskeleton of the cell – the exact same actors that Yersinia targets during infection to stay outside the macrophages.
Later during infection, cell death and inflammation play a major role in controlling infections, since macrophages often kill themselves in a manner that alerts other immune cells in a process called pyroptosis. The second interaction points that we investigate is related to that process: How easily can the inflammatory signaling that is required to alert other cells be activated? We identify that a chronic strain of Adenovirus (mAdV3), which on its own causes neither inflammation nor cell death, decreases the responsiveness of pyroptosis-inducing signaling hubs – the inflammasomes. More specifically, inflammasomes that rely on an adaptor protein called ASC were much less responsive to secondary stimuli after infection with mAdV3. This is highly meaningful, since several of the bacterial pathogens we investigate normally trigger an activation of ASC-dependent inflammasomes, but with mAdV3 dampening the responsiveness, there are severe impacts on how easily secondary infections can be communicated to neighboring cells after macrophages were infected.
In summary, we set out to explore the vastly open space of co-infections on a molecular level. We wanted to know if there are general trends for co-occurring pathogens and tried to characterize molecular mechanisms that cause these. While this study provides a proof of principle, there is much more to be done: Other pathogens, host model systems, readouts and setups will have to be explored in the future to further strengthen our ability to understand how pathogens interact and cause the increased severity of co-infections observed in clinical studies. Ultimately, we hope that the mechanistic knowledge that we uncovered can also serve to better understand the strategies employed by pathogens, as well as how our cells respond to microbial invaders, thereby leading to new anti-infective treatments.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Ask the Editor - Immunology, Pathogenesis, Inflammation and Innate Immunity
Got a question for the editor about the complement system in health and disease? Ask it here!
Continue reading announcementRelated Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in