What does control the biodiversity of the Coral Triangle?
Published in Ecology & Evolution

Anthropogenic stressors have a considerable influence on marine ecosystems. Human activities such as fishing, coastal development and agriculture, together with global climate warming threaten ocean biodiversity and ecosystem functioning. The Coral Triangle is the most diverse and biologically complex marine ecosystem, called home by over 75% of known species of reef-building corals, 3,000 species of reef fish and 75% of known mollusks.
In the past 30 years, ocean temperatures around the CT have risen by approximately 0.1 °C per decade and are expected to climb an additional 1-4 °C by 2100. An increase of more than 2 °C will eliminate most coral-dominated reefs, with the potential of impacting the livelihoods of 120 million people and cutting protein supplies to one billion people. Corals are foundational: If they disappear, marine biodiversity as a whole will reduce greatly.
Conservation management and the planning or adoption of mitigation strategies in any ocean realm requires information about the stressors, and most importantly about the distinct ecoregions that demark unique assemblages of species. The potential of a marine protected area to retain biodiversity and restock species abundance beyond its border, is inherently linked to some form of regional discretization and population connectivity, such as the degree and directionality of larval dispersal across ecoregions. In most of the world Ocean, and in the CT, defining the ecoregions is complicated by data sparseness and by the large-scale, time-dependent dispersal of ocean currents. Indeed, ocean currents and water characteristics change in space and time, and predictions of community susceptibility to these changes remain elusive.
This knowledge is especially important when biodiversity must rebuild following a devastating damage, as for example following bleaching events in the CT, because ecoregions and connectivity contribute to colonization and resilience of a given reef. In most of the world’s Oceans, and in the CT, defining ecoregions and connectivity is complicated by data sparseness and by large-scale, time-dependent ocean currents. Direct observations of species distributions are indeed sparse in space and time, concealing the relationships between ecoregions, climate variability and ocean currents. Model simulations of ocean circulation and larval transport help, but they are limited in their spatial and temporal resolution.
Our work provides a pathway towards filling this fundamental knowledge gap in coral ecology introducing an unsupervised machine learning (ML) approach that exploits the dynamical relationship between sea surface temperature anomalies (SSTa) and sea surface height anomalies, and therefore currents, at spatiotemporal frequencies pertinent marine communities at latitudes comprised between 45oN and 45oS. The ML framework, schematically summarized in Fig. 1, is applied to the SSTa from a high resolution reanalysis, GLORYS, from 1993 to 2017. SSHa are grouped according to the El Niño Southern Oscillation (ENSO) phase, given the strong impact of ENSO on current directionality in the CT area.
Fig. 1 Schematic of the ML workflow proposed in the paper.
The results of our analysis indicate that the extraordinary biodiversity that we observe today is maintained and enhanced by ENSO variability (Figure 2). While ENSO has long been associated with coral reefs mortality, due to the prolonged higher-than-normal temperatures experienced in some areas during its positive and negative phases, ENSO appears to be highly beneficial to biodiversity. It enhances the large-scale exchange of genetic material between the Indian Ocean and the CT during La Niña years, and between the CT and the area to the east in neutral conditions. Most importantly, we provide, a climatic context to interpret the evolution of species richness in the Indo-Pacific through geological times.
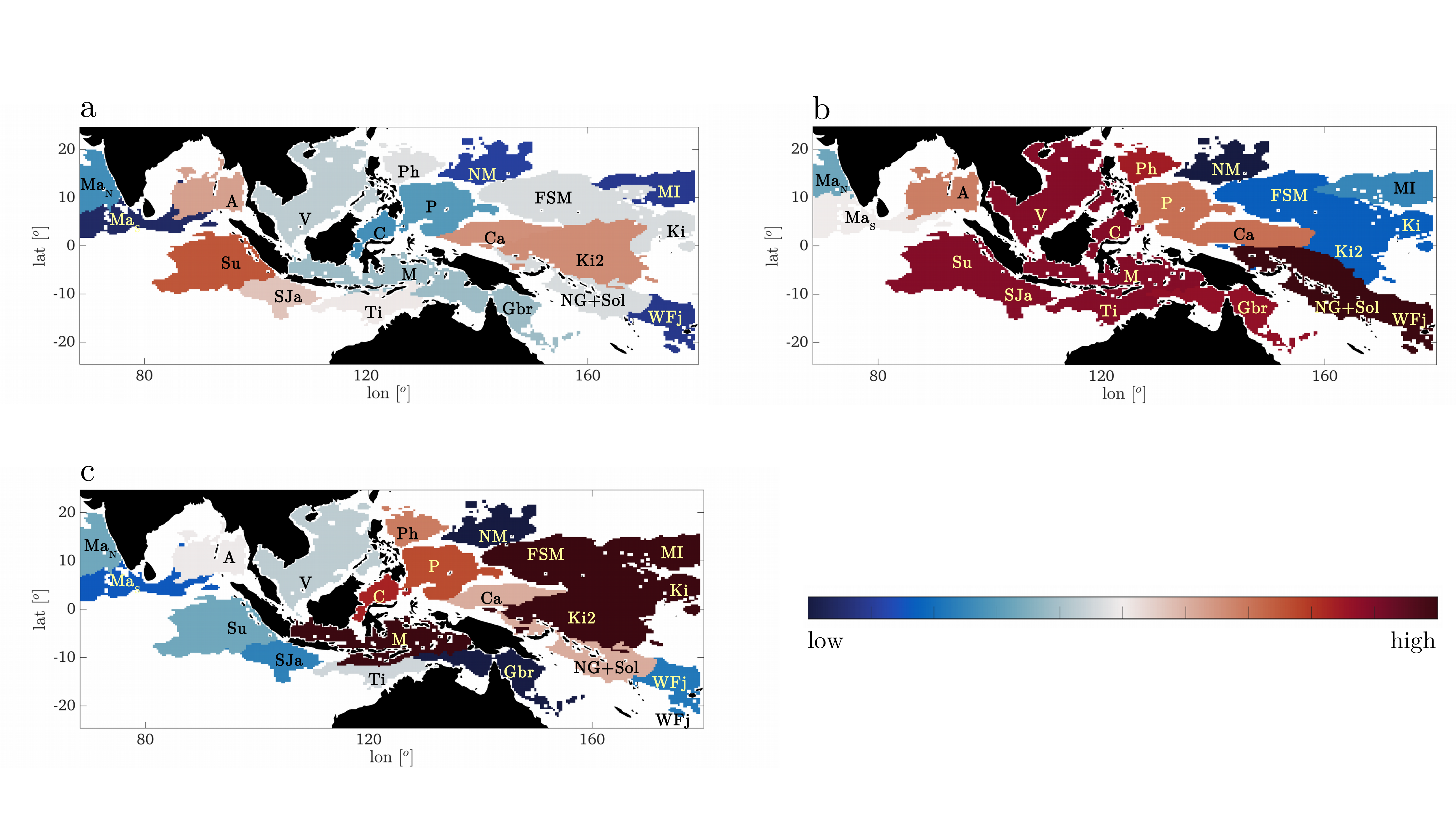
Fig. 2. Biodiversity Score over the period 1993-2017 for the domains found during (a) El Niño years, (b) La Niña years, (c) Neutral years. The color scale is linear (low = 0.03, high = 0.08).
The ecoregion identification and biodiversity score we propose taken together with the recovery potential metric introduced in this work, can identify appropriate temporal and spatial windows where connectivity-based restoration efforts and monitoring should be prioritized. This component is relevant to regional, industry and community groups for which climate adaptation and biodiversity conservation is particularly important. Given the societal relevance of reef ecosystems, the potential of the framework presented is profound.
Follow the Topic
-
Communications Biology

An open access journal from Nature Portfolio publishing high-quality research, reviews and commentary in all areas of the biological sciences, representing significant advances and bringing new biological insight to a specialized area of research.
Related Collections
With Collections, you can get published faster and increase your visibility.
From RNA Detection to Molecular Mechanisms
Publishing Model: Open Access
Deadline: May 05, 2026
Signalling Pathways of Innate Immunity
Publishing Model: Hybrid
Deadline: May 31, 2026

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in