What if you could take your lab in a backpack to explore life on Earth?
Published in Protocols & Methods
Studying the diversity of life on our planet is no small task. Millions of species remain to be discovered or described in detail, while the loss of natural habitats and biodiversity continues to accelerate. Conservation and biosurveillance-focused groups are faced with the immense task of characterizing baseline biodiversity data, documenting how biological communities change and detecting invasive pests and pathogens before they become harmful. To do so, access to the appropriate scientific tools is are required.

DNA sequencing has become one such tool for biologists to understand life on our planet. Recent advancements in sequencing technology and decreases in cost have inspired global initiatives such as the Earth BioGenome Project, which aims to sequence the genomes of all of Earth's eukaryotic biodiversity. However, there still remains a great divide between where biodiversity is concentrated (primarily in tropical regions of the world) and where researchers have access to DNA sequencing infrastructure (typically in wealthy nations). Collecting biological samples and then transporting them abroad for analysis has been the status quo, but this often increases the time it takes to generate, analyze and report on the biological information, and can contribute to biases in global representation of biodiversity as well as scientific capacity building.

Examples of reptile and amphibian diversity in Ecuador. From top left to bottom right: Amazon tree boa, rain frog, Pinocchio lizard, Galapagos land iguana, fringed leaf tree frog, tropical salamander (Photos by Aaron Pomerantz).
The ability to sequence biological samples within the country of origin with portable, inexpensive laboratory equipment can bring major benefits for biodiversity monitoring and explorations, while simultaneously creating opportunities for developing local scientific capacity. Additionally, using DNA sequencing to rapidly characterize invasive pests and pathogens at or near the site of detection can mitigate their negative impacts on health, ecosystems and economies.
Miniaturized scientific instruments, such as portable thermocyclers and nanopore-based nucleic acid sequencing devices, have emerged as powerful tools for biologists, as they:
-
are relatively inexpensive compared with traditional, bulky molecular laboratory equipment
-
allow for in situ processing of genetic material,
- offer opportunities to empower local scientists and conservation agencies that currently rely on international research facilities.
At the heart of advancements in miniaturized genomics equipment is the small MinION sequencing device (developed by Oxford Nanopore Technologies), which conducts ‘nanopore sequencing’. This process occurs via changes in an electric current that are measured and characterized when a DNA molecule is funneled through a biological nanopore.
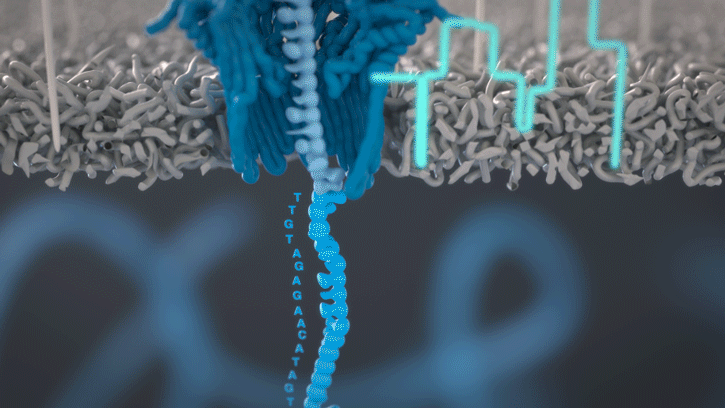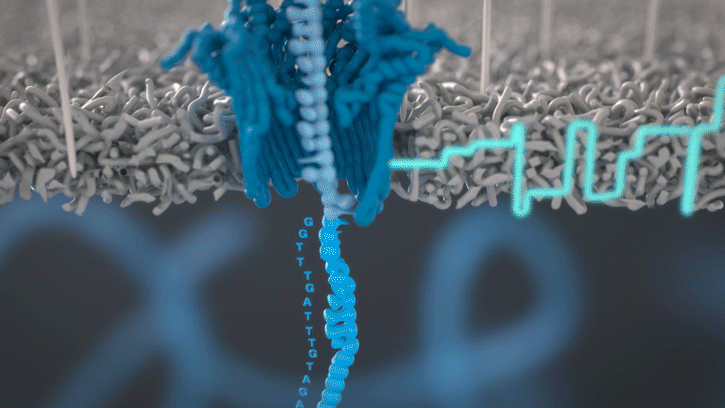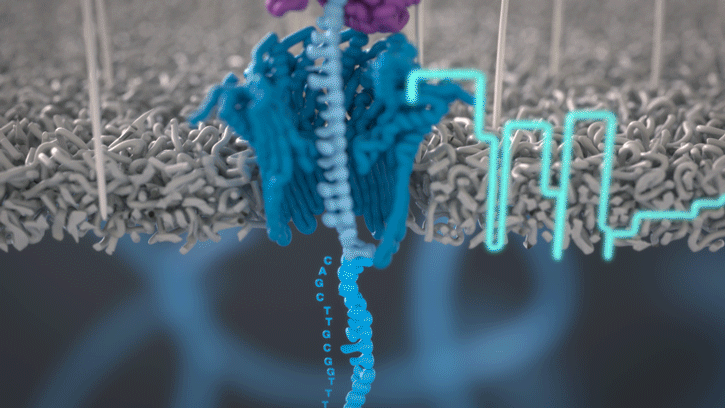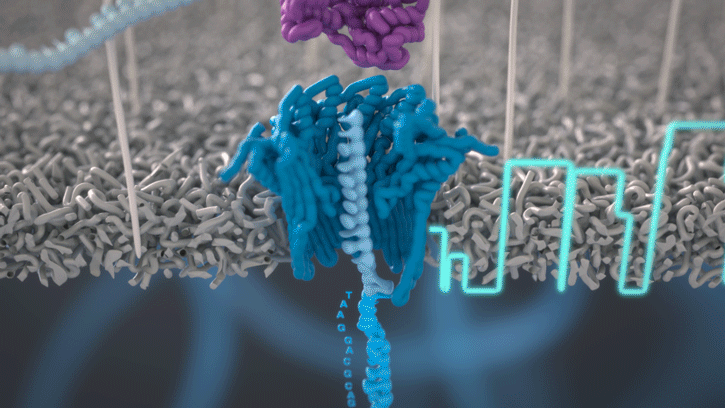
Animation of nanopore sequencing. The nanopore (blue) is embedded in an electro-resistant membrane (gray). Each nanopore corresponds to its own electrode connected to a channel and sensor chip, which measures the electric current that flows through the nanopore. When a molecule passes through a nanopore, the current is disrupted to produce a characteristic ‘squiggle’. The squiggle is then decoded using basecalling algorithms to determine the DNA sequence in real time. [Oxford Nanopore Technologies]
Over the past few years, portable genomics laboratories have been deployed around the world in a diverse set of ecosystems and settings, including the rainforests of the Congo, Ecuador and Madagascar, out at sea and on ice caps, as well as being used to monitor disease outbreaks such as Ebola in West Africa or Zika in Brazil, and for educational programs at universities or in the field. Following several years of experimentation and refinement of MinION-based methods by the scientific community, combined with the optimization of other commercially available field-deployable laboratory equipment, we felt that there was now an opportunity to put together a step-by-step protocol for DNA amplicon sequencing (a technique that amplifies a short piece of DNA which can subsequently be used for species identification purposes) using miniaturized laboratory equipment, as a resource for the community and new users interested in getting started.

As a contribution to Nature Protocols, we consolidated currently available best practices for performing amplicon sequencing experiments outside of conventional laboratory environments based on research groups who have processed samples and applied portable genomics tools under field conditions (see Pomerantz et al. 2022). Additionally, we provided cost-effective strategies for sequencing a high number of samples and presented a simplified downstream bioinformatics workflow to generate highly accurate DNA sequences, which can then be used to search against databases for identifying the specimen of interest.
We hope this protocol serves as a useful resource for others to get started with assembling their own portable genomics labs and empowers local researchers in particular. The approach outlined in this paper can be adapted for various projects that aim to perform real-time DNA amplicon sequencing, such as within-country biodiversity assessment, wildlife forensic efforts, on-site invasive species detection, and educational genomics programs, as users can tailor the protocol to work with any taxa of interest in a relatively time- and cost-efficient manner.
For an open-access, read only link use: https://rdcu.be/cK7hP
Reference for the Nature Protocols paper:
Follow the Topic
-
Nature Protocols

This journal publishes secondary research articles and covers new techniques and technologies, as well as established methods, used in all fields of the biological, chemical and clinical sciences.



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in