When a Natural Compound Disrupts an Alzheimer’s Protein
Published in Mathematics
Exploring How a Natural Compound May Disrupt a Key Alzheimer’s Protein
Alzheimer’s disease is often discussed in terms of memory loss and cognitive decline, but at its core, it begins with changes at the molecular level. One of the main contributors to the disease is a small protein called amyloid-beta. When amyloid-beta molecules stick together, they form toxic clusters that interfere with normal brain function.
This research set out to explore a simple question: Can natural compounds interfere with the formation or stability of these harmful protein clusters?
Looking to Nature for Clues
Natural products have inspired many modern medicines, and compounds from Panax ginseng have long been associated with brain health. These compounds, called ginsenosides, are widely studied for their biological activity and relatively low toxicity.
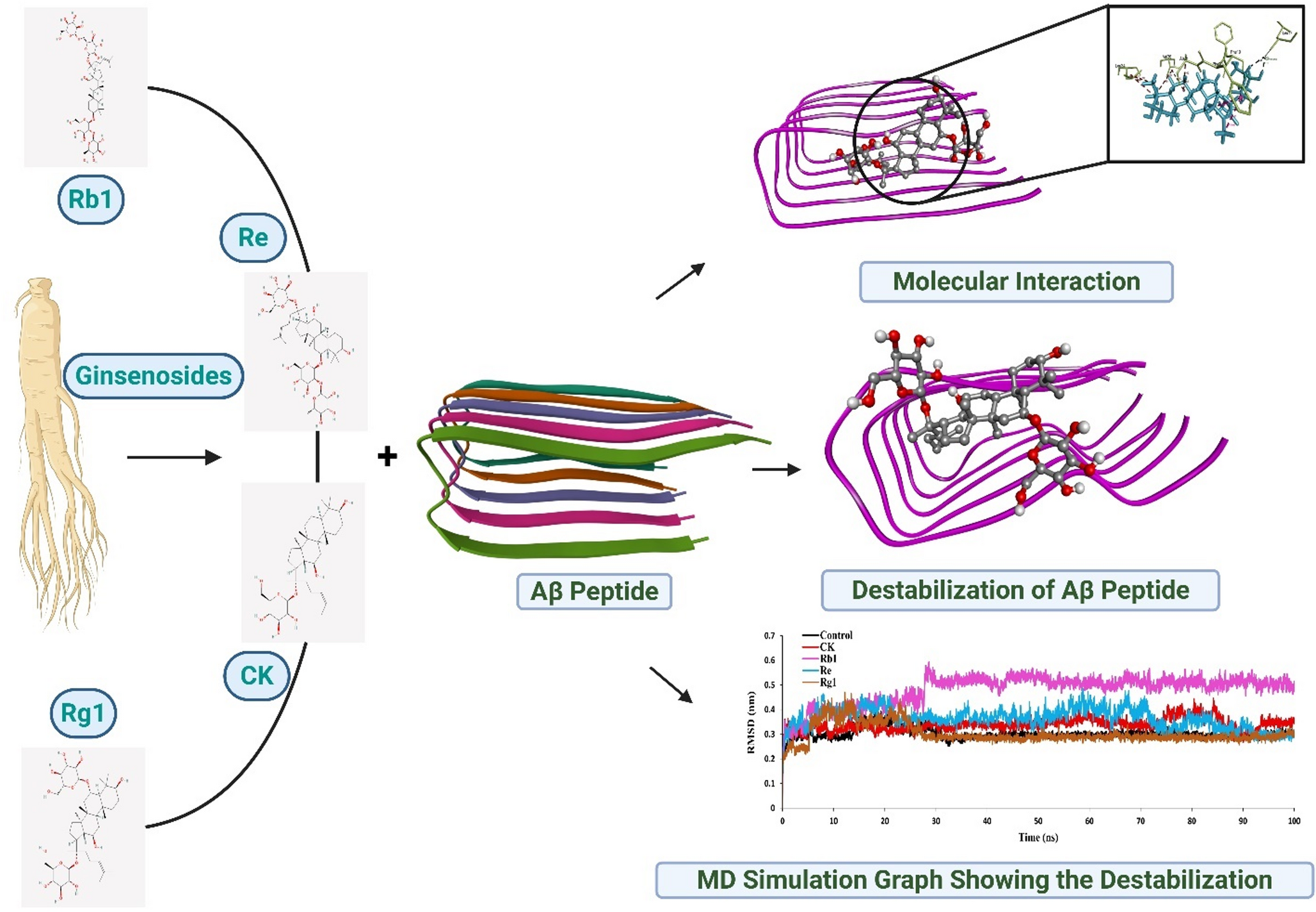
In this study, four well-known ginsenosides—Compound K, Rb1, Re, and Rg1—were examined to see how they interact with amyloid-beta protein clusters. Instead of starting in the lab, the work used computer-based methods that allow researchers to observe molecular interactions in detail.
Figure 1: Visualizing the Molecular Battle Against Alzheimer’s
Watching Molecules in Motion
Using molecular docking, the researchers first tested how well each ginsenoside could attach to amyloid-beta clusters. All four showed promising binding, suggesting they could interact with the protein.
The next step was to observe what happened over time. Molecular dynamics simulations—essentially virtual “movies” at the atomic level—were used to see whether these compounds could actually disturb the structure of amyloid-beta clusters.
One Compound Stands Out
Among the four compounds, ginsenoside Rb1 consistently showed the strongest effect. In the simulations, amyloid-beta clusters became less stable and more disorganized when Rb1 was present. This suggests that Rb1 may weaken the interactions that stabilize these harmful protein structures.
The other compounds showed milder effects, and one caused partial disruption—but none matched the consistent impact of Rb1.
Why This Matters
This study does not claim to offer a treatment for Alzheimer’s disease. Instead, it provides early-stage insight into how natural compounds might influence disease-related proteins at the molecular level.
By identifying ginsenoside Rb1 as a particularly promising candidate, the research helps guide future laboratory and experimental studies. It also highlights the value of computational tools in narrowing down potential therapeutic leads before moving to more complex and costly experiments.
While the work happens quietly on computers rather than in visible experiments, it contributes an important piece to the larger effort to understand—and eventually address—Alzheimer’s disease.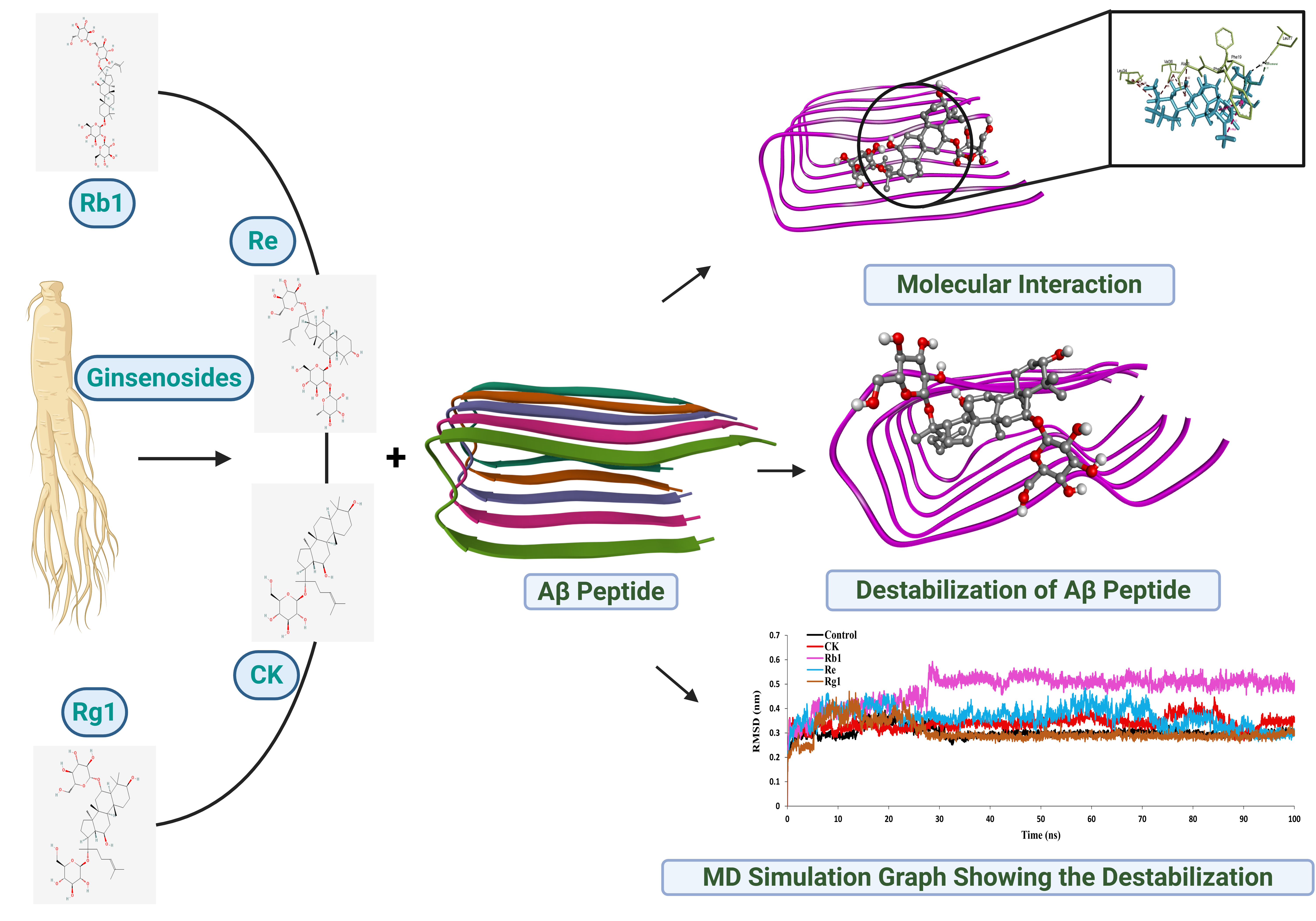




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in