When cells change shape: how organelle dynamics led us to a new way of scoring EMT
Published in Protocols & Methods, Cell & Molecular Biology, and Computational Sciences
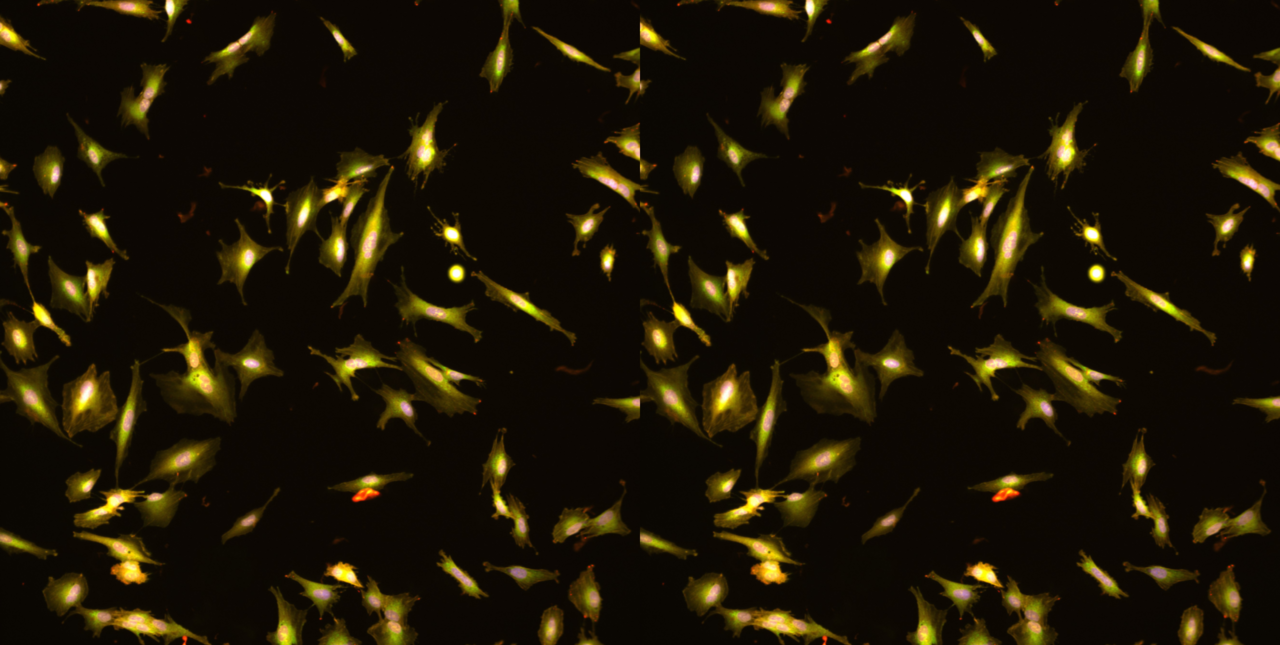
The beginning
Back in early 2023, we were wrestling with a familiar problem in EMT research: the data is barely consistent. Western blot results shift unpredictably, and every apparent pattern raises new questions. EMT felt less like a clean biological program and more like trying to sketch the outline of something that refused to hold still.
Around that time, our supervisors, Jonas and Malgorzata, came to us with an idea they had been carrying around for a while. “If the cytoskeleton of certain cell lines changes so dramatically during EMT, what happens to the organelles? Could those internal changes tell us where a cell belongs along the EMT spectrum?”
It was the kind of question that sounds simple at first but becomes more interesting the longer you think about it. Cellular morphology clearly changes in microscopy images; anyone who has looked at cells long enough through a simple brightfield microscope knows they rarely keep the same shape for long. But the idea of using those internal changes to quantify EMT was something new.
The suggestion came from a real place of frustration. After months of chasing inconsistent protein and transcript readouts, we were eager to find a complementary way to track this notoriously plastic process. Maybe organelles carried a signature that molecular markers alone could not give us.
So, we decided to test it.
From a Few Images to a Full High-Throughput Project
The start of the project was almost deceptively simple. We took an easy-to-grow cell line, stained the organelles, and took a few images with a confocal microscope. No automation, no special machinery, just enough to see whether organelles behaved differently when cells were pushed into EMT.
Even those first images were surprising. The mitochondria looked more granular; the ER texture shifted, and the nuclei stretched in unfamiliar ways. It was not subtle. It was as if EMT had left fingerprints all over the inside of the cell.
Encouraged, we scaled up.
What began as a handful of images turned into high-content imaging across multiple plates, with thousands of wells and eventually hundreds of thousands of cells. Staining six organelles at once turned out to be a technical art form. Some dyes were overexposed, and others refused to stain evenly. And then there was the microscopy problem: the high-throughput machine at the core facility was often booked. Imaging happened whenever time was available, usually late at night, early mornings, weekends, and occasionally sandwiched between someone else’s bookings.
But every dataset we collected showed clearer and clearer signs that the organelles were indeed reorganizing in systematic ways. Those irregular imaging hours started to feel worth it.
When Biology Becomes Data Science
At some point, the biological part of the project gave way to something else entirely. Each cell produced 2,608 morphological features. Multiply that by hundreds of thousands of cells, and suddenly, we gathered a respectable dataset. We realized we could no longer understand this project with standard tools, which meant learning Python, machine learning tools, and computational workflows from scratch.
To be honest, neither Francesca nor I had ever written serious code before. We were biologists who liked nice images, not data scientists. Our first scripts were… functional. They ran, but slowly.
Still, they taught us a lot. The first simple classifiers showed hints of clusters that lined up with EMT time points. That was the first moment we thought, “This might actually be working.”
Then Benjamin joined the project, turning the improvised scripts into something much more stable. The analysis pipeline became clearer, more reproducible, and far more reliable.
For the first time, the data started speaking a coherent language.
The Real Turning Point
The moment that changed everything came when we compared our morphology-based scores to EMT progression derived from RNA-seq. We expected a rough trend — maybe early and late stages aligning somewhat.
Instead, the patterns were nearly identical.
It was a surreal moment. After months of troubleshooting stains, debugging scripts, and trying to believe that the organelles were telling us something real, the RNA-seq comparison showed a near one-to-one correspondence. That was when the project crossed the line from “interesting idea” to “this might actually be a new way to look at EMT scoring.”
And then came another key idea, thanks to Wenyang, to turn our classifier probabilities into a continuous EMT score. Suddenly, the output was not just categories; it was a smooth trajectory that captured hybrid states and reversal dynamics much more naturally. Everything became more intuitive.
Building a Paper Out of Trial, Error, and Persistence
Of course, what ends up in a manuscript is only the polished surface. Behind it were many less glamorous moments: staining failures, segmentation errors, code that ran for hours only to produce an empty file, and repeated experiments during revision, including experiments done abroad, reassuring us that what we saw was real and reproducible.
But that process was also the most rewarding part. With each iteration, the method became stronger. And slowly, a manuscript emerged: not just a collection of results, but a story of problem-solving, learning new skills, and watching a small idea turn into something much larger than we expected.
The Bigger Picture
What began as a speculative question became a framework that helped us secure funding for a large-scale drug screening and opened a new way to score epithelial–mesenchymal plasticity. It also showed us how much biological information is hidden in structures we often overlook. Organelle morphology turned out to carry a remarkably rich signature of cellular identity.
Looking back, the journey was shaped by curiosity, persistence, and a willingness to step outside our comfort zone. We learned new methods not because we wanted to, but because the project demanded it. And in doing so, we discovered a new perspective on EMT, one that connects imaging, computation, and cell morphology in a way we could not have predicted at the start.
Follow the Topic
-
Communications Biology

An open access journal from Nature Portfolio publishing high-quality research, reviews and commentary in all areas of the biological sciences, representing significant advances and bringing new biological insight to a specialized area of research.
Related Collections
With Collections, you can get published faster and increase your visibility.
Lipids in Cell Biology
Publishing Model: Open Access
Deadline: Mar 03, 2026
Forces in Cell Biology
Publishing Model: Open Access
Deadline: Apr 30, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in