When Modifications Rewrite Networks: How PTMs Reprogram Chaperones into Epichaperomes
Published in Neuroscience, Physics, and Protocols & Methods
A new way of thinking about post-translational modifications
Post-translational modifications (PTMs) are often described as molecular fine-tuners—chemical tags that adjust a protein’s activity, localization, or stability. Yet, in our recent TIBS review, we propose that PTMs do far more than modulate single-protein behavior. They can act as molecular encoders that direct proteins toward distinct structural and functional states, effectively writing a code that governs how proteins assemble into higher-order architectures.
This concept takes on particular importance for the cellular chaperone system—long known for its role in protein quality control. We argue that PTMs on chaperones and co-chaperones can reprogram these proteins into stable, supramolecular assemblies called epichaperomes. These assemblies do not fold clients; instead, they serve as scaffolds that reorganize protein-protein interaction (PPI) networks, thereby reshaping cellular pathways at a systems level.
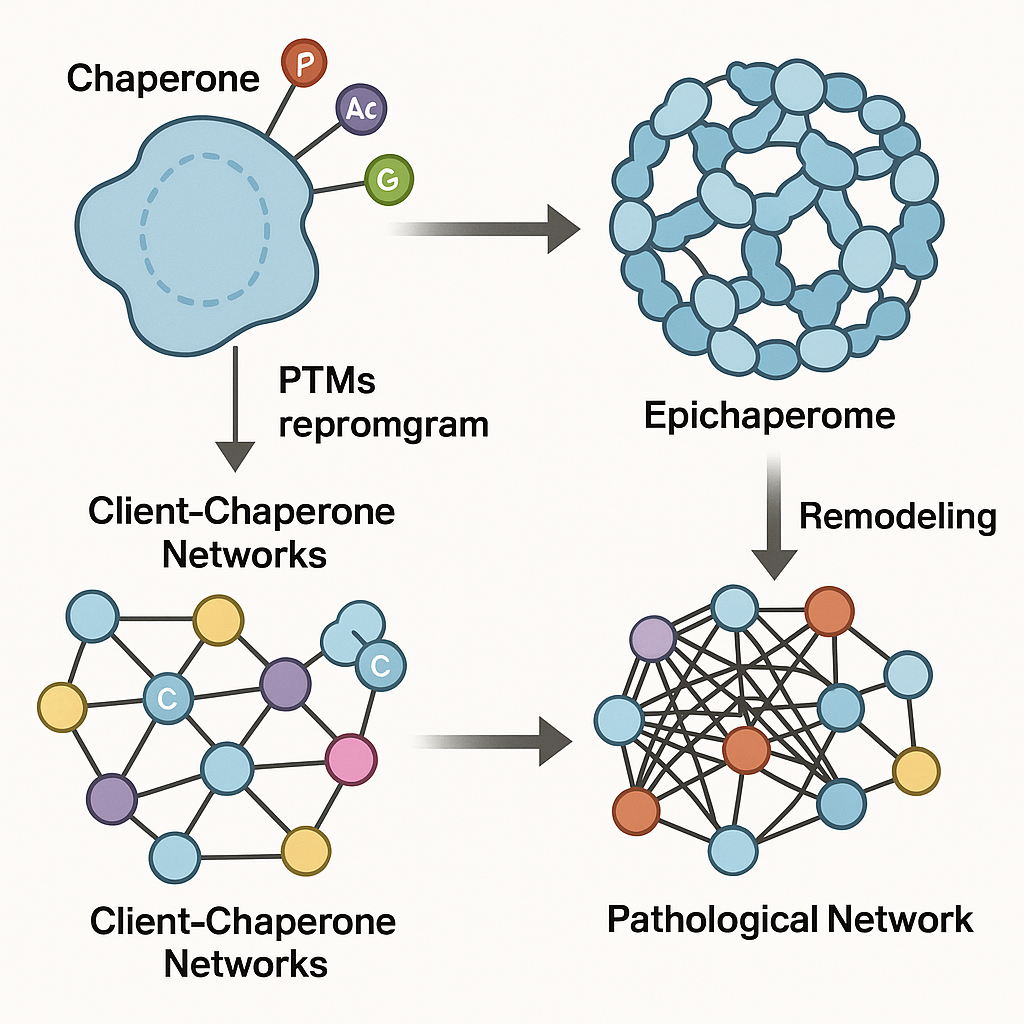
From helpers to organizers: the emergence of epichaperomes
In healthy cells, chaperones form dynamic complexes that transiently assist thousands of proteins through folding, maturation, or degradation. This activity is rapid and reversible, ensuring proteome flexibility.
Under stress or disease conditions, however, PTMs—such as phosphorylation, acetylation, or glycosylation—can shift chaperones into structurally distinct states that favor prolonged, multivalent interactions. These modified chaperones coalesce into network-like assemblies, or epichaperomes, that scaffold large sets of proteins into coordinated modules.
Rather than serving the folding cycle, these assemblies repurpose chaperones as network organizers, allowing the cell to rapidly rewire pathways required for survival under adverse conditions. The result is a proteome that is reorganized—not by changes in gene expression, but by a re-encoded interaction architecture.
PTMs as encoders of structural and functional state
Many PTMs occur within intrinsically disordered regions (IDRs)—flexible protein segments that can adopt multiple conformations. These regions act as sensitive molecular switches. A single phosphorylation or glycosylation event can reshape an IDR’s charge, dynamics, and binding preferences, influencing how it engages partners.
In the context of chaperones such as HSP90, HSC70, or GRP94, PTMs within IDRs can stabilize specific conformations that promote the multimerization and interlocking of chaperone units. In this way, PTMs encode assembly state and network participation, determining whether a chaperone operates in its canonical, dynamic mode or as part of a stable epichaperome scaffold.
Epichaperomes: from molecular code to cellular consequence
Once formed, epichaperomes impose structure on the cellular interactome. By sequestering and redistributing proteins, they create new PPI topologies that underlie the reprogramming of signaling, metabolism, and stress-response networks.
In cancer, epichaperomes stabilize pathways that sustain high proliferation and survival. In neurodegenerative disease, they can propagate dysfunction by trapping neuronal and glial proteins into maladaptive assemblies. Across these diverse conditions, the unifying feature is network control—the ability of epichaperomes to coordinate complex cellular behavior through scaffolding rather than expression changes.
Therapeutic insight: targeting architecture, not abundance
Traditional therapeutic strategies often aim to inhibit or degrade specific proteins. The epichaperome framework suggests a complementary approach: targeting the architecture that connects them.
Small-molecule disruptors such as PU-H71 and related compounds selectively bind to chaperones when they are incorporated into epichaperomes, disassembling the scaffold without affecting normal chaperone function. This network-selective mode of action provides a precision route to normalize dysregulated interactomes—an approach already advancing into clinical testing for cancer and neurodegeneration.
A broader perspective on protein regulation
The concept that PTMs can encode supramolecular assembly opens a new dimension in cell biology. It reframes the proteome not as a static list of proteins and modifications but as a dynamic, PTM-programmed network of assemblies that defines cell state.
Understanding this encoding system will require integrating proteomics, structural biology, and systems modeling to decipher how specific PTM combinations dictate when and where epichaperomes form. As such, PTMs are not merely chemical decorations but molecular instructions that can reconfigure entire biological networks.
Looking ahead
Our hope is that this framework encourages scientists to view PTMs not only as regulators of single-protein function but as architectural determinants of cellular organization. By decoding the PTM-driven language of epichaperomes, we can begin to understand—and perhaps control—how cells reshape their networks in health and disease.
Read the full open-access article:
Chu F., Sharma S., Ginsberg S.D., Chiosis G. (2025). PTMs as molecular encoders: reprogramming chaperones into epichaperomes for network control in disease. Trends in Biochemical Sciences, 50(10): 892–905.
DOI: 10.1016/j.tibs.2025.07.006

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in