Why cultivation still matters in microbiology
Published in Ecology & Evolution and Microbiology

When scientists talk about the “microbial dark matter” or the “uncultivated microbial majority”, they refer to the overwhelming majority of microbes that we know exist (because we can detect them in metagenomes or 16S rRNA gene amplicon studies) but that so far have not been grown in the lab. Despite decades of research, most environmental microbes remain uncultivated. This limits what we can truly learn about them: their physiology, their interactions, and their ecological importance.
In our new paper in Nature Communications, we set out to bring some of this uncultivated microbial majority to the lab. Using high-throughput dilution-to-extinction cultivation with carefully designed artificial media, we isolated 627 axenic strains of freshwater microbes from 14 Central European lakes. The retrieved diversity includes many of the most abundant freshwater genera that are notoriously underrepresented in public culture collections and many taxa which had never been grown before. Our collection includes strains from 15 of the 30 most abundant freshwater bacterial genera, representing up to 72% of genera detected in the original samples (average 40%) and encompassing key lineages widespread across global freshwater systems.
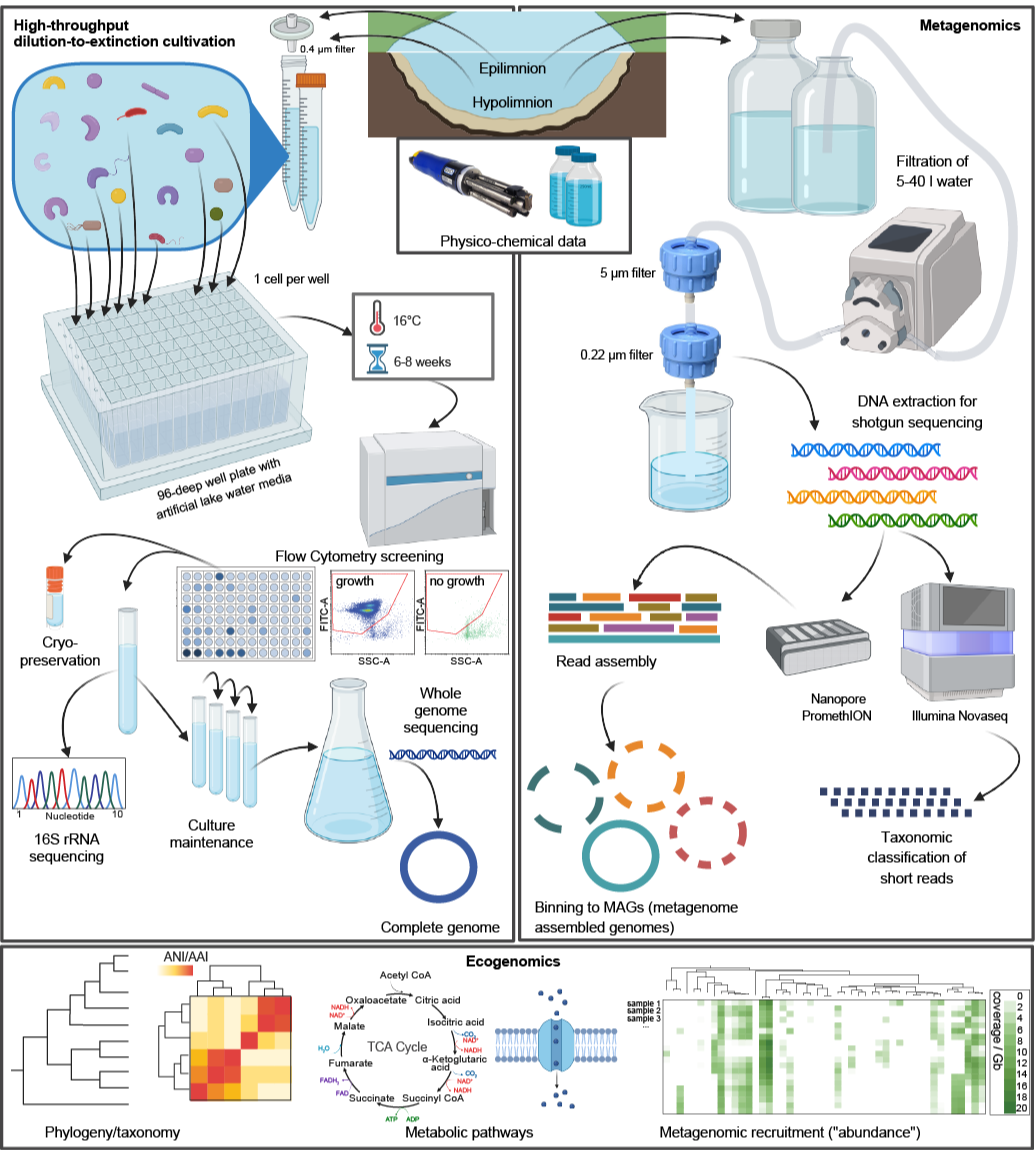
Workflow for the isolation of aquatic bacteria (left side), metagenomic sequencing (right side), and ecogenomic analyses (bottom). Source: Fig. 1 from Salcher et al. (2025) Nature Communications 16, 7971.
Why genomes are not enough
Over the last 15 years, metagenomics has transformed microbiology. We can now reconstruct genomes (metagenome assembled genomes, i.e. MAGs) directly from environmental DNA, reconstruct metabolic pathways, and infer evolutionary histories without ever having a culture in hand. These approaches are incredibly powerful, but they also have limitations. Metagenomes provide snapshots of metabolic potential, i.e., tell us what a microbe could do, but not whether or when it actually does it under natural conditions. Further, many genes are poorly annotated or have unknown functions. And without cultivated representatives, we can’t test hypotheses experimentally, we can’t measure growth, metabolism, or physiological limits. Moreover, only axenic cultures enable us to study strain-level microdiversity1, as MAGs are always a composite assembly of many closely related strains that coexist in nature.
Cultivation is thus still essential for closing the loop between genes and functions. It allows us to conduct experiments that validate predictions from genomic data and refine models2. And just as importantly, it gives us physical reference strains that other researchers can work with worldwide.

Mondsee (left) and Attersee (right; seen from the top of Schafberg, Austria), two of the lakes from where we isolated many new microbes. Image credit: M. Salcher
How to grow “unculturable” microbes
Why are so many microbes uncultured? In part because traditional microbiology methods are biased toward fast-growing species that thrive in nutrient-rich conditions, organisms we call “copiotrophs.” But in natural environments like lakes and oceans, many dominant microbes are oligotrophs: slow-growing organisms adapted to extremely low nutrient concentrations. These oligotrophs don’t grow in nutrient-rich media or on agar plates (Why would they? In nature they are free-living and planktonic), besides other unknown growth requirements.
The secret to growing oligotrophs is simple in principle but tricky in practice: we must recreate their natural environment. In our study, we used dilution-to-extinction techniques with defined media that closely mimic lake water chemistry. By lowering nutrient concentrations and isolating single cells in multi-well plates, we reduced competition and allowed slow growers the chance to thrive. This approach worked remarkably well. We recovered strains belonging to the most abundant freshwater genera (e.g., the freshwater SAR11 genus Fontibacterium, Planktophila, Methylopumilus) and many which were known only by alphanumerical placeholder names (e.g., acIV, MWHUniP1)3,4.
So why do not more scientists use dilution-to-extinction cultivation? Well, it’s a long-term process and still very laborious even with semi-automated workflows like a flow cytometer with a 96-well plate loader. We screened more than 6’000 wells and did more than 1’200 PCRs to end up with ~600 pure cultures. Of all these cultures, still, a large part was copiotrophs and not the targeted abundant oligotrophs, and we also obtained the same genera over and over again. So in the end, it’s a numbers game, the more you isolate, the higher the chances for cultivating something really interesting. Further, cultivation is slow business: it took us at least 6-10 months to obtain genomes, as screening and upscaling of these slow growers takes time. MAGs, on the other hand, can be sequenced and binned almost immediately after sampling. Last but not least, cultivation work is tedious with a high risk of contaminating or loosing cultures as sometimes strains stop growing for unknown reasons after a few transfers and can’t be revived from cryostocks.

Left: Preparation for dilution-to-extinction isolation with 96-well plates filled with artificial lake water medium. Right: Part of the culture collection grown in 15 ml tubes. Image credit: M. Salcher
What we found - the bigger picture
We genome-sequenced 87 strains of our culture collection and described two new families, nine new genera, and 41 new species. Many had the hallmarks of oligotrophic lifestyles with streamlined genomes. Genome sequencing further revealed a high diversity of lifestyles and metabolic pathways.
Perhaps the most important outcome of our work is the culture collection itself. These strains are now available for further studies as model organisms to test ecological theories, study microbial interactions, and explore functional roles in biogeochemical cycles. In doing so, we not only expand our understanding of microbial diversity but also build the experimental foundation needed to address pressing ecological questions from how lakes cycle carbon and nutrients to how microbial communities respond to climate change.
By cultivating organisms that are both abundant and ecologically relevant, we hope to enable a new wave of experimental freshwater microbiology. Just as the marine SAR11 clade (Pelagibacter) became a cornerstone of marine microbiology once it was cultured5, our strains may now serve as reference systems for freshwater lakes worldwide. Cultivation is often seen as “old-fashioned microbiology.” But as this work shows, it remains an essential complement to modern genomics. By bridging the gap between DNA data and living cells, we can move from what is out there to how it works.
References:
1. Layoun et al. (2024) Flexible genomic island conservation across freshwater and marine Methylophilaceae. ISME J. 18, wrad036. https://doi.org:10.1093/ismejo/wrad036
2. Fernandes et al. (2025) Ecophysiology and global dispersal of the freshwater SAR11-IIIb genus Fontibacterium. Nat. Microbiol. https://doi.org:10.1038/s41564-025-02091-8
3. Newton et al. (2011) A guide to the natural history of freshwater lake bacteria. Microbiol. Mol. Biol. R. 75, 14-49. https://doi.org:10.1128/mmbr.00028-10
4. Chiriac, Haber, & Salcher (2023) Adaptive genetic traits in pelagic freshwater microbes. Environ. Microbiol. 25, 606-641. https://doi.org:https://doi.org/10.1111/1462-2920.16313
5. Giovannoni (2017) SAR11 bacteria: The most abundant plankton in the oceans. Ann. Rev. Mar. Sci. 9, 231-255. https://doi.org:doi:10.1146/annurev-marine-010814-015934
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in