Teaming up to investigate the landscape of antimicrobial resistance mutations in fungi
Published in Microbiology and Protocols & Methods

The climb to the landscape view
Infections caused by fungal pathogens are a serious health threat and socio-economic burden. In clinics, fungal infections are becoming more frequent, while in agriculture, fungal diseases cause major crop losses. To treat infections and to prevent fungal infections, we rely on antifungal drugs from different classes. However, antimicrobial resistance (AMR) is threatening our ability to control these pathogens. The widespread use of antifungals has driven the emergence of resistant strains and populations unresponsive to treatments1.
When I first started working on antifungal resistance as an undergraduate student in 2020, I quickly stumbled across a problem that slowed me down: data on resistance mutations were scattered across the scientific literature. The MARDy database2 was available at that time, but it was unfortunately incomplete. I remember thinking that having a comprehensive, centralized database of resistance mutations would be extremely useful (and that sentiment was definitely shared). Without complete, organized and accessible data, it’s harder to design meaningful research on the subject, to monitor resistance in populations, to adjust treatments when resistance emerges or to explore ways to bypass resistance mechanisms.
In 2023, our lab leader Christian Landry initiated the curation of scientific papers reporting AMR mutations in fungi. The goal was to build a dataset of all known drug resistance mutations in human and plant fungal pathogens to give scientists easy access to the data. As the instigator of the Canadian training program on the evolution of fungal pathogens (EvoFunPath), he also brought together graduate students, postdoctoral fellows and professors from across Canada and Europe to help with the work. The initial plan was to review all the papers together over a few intense days with some of us meeting in person at Université Laval and others joining online. However, we quickly realized that we had underestimated both the sheer volume of data in the literature and the complexity of curating it carefully. Therefore, what we thought would be an easy walk in the park turned into a long and difficult hike to the top of a mountain.
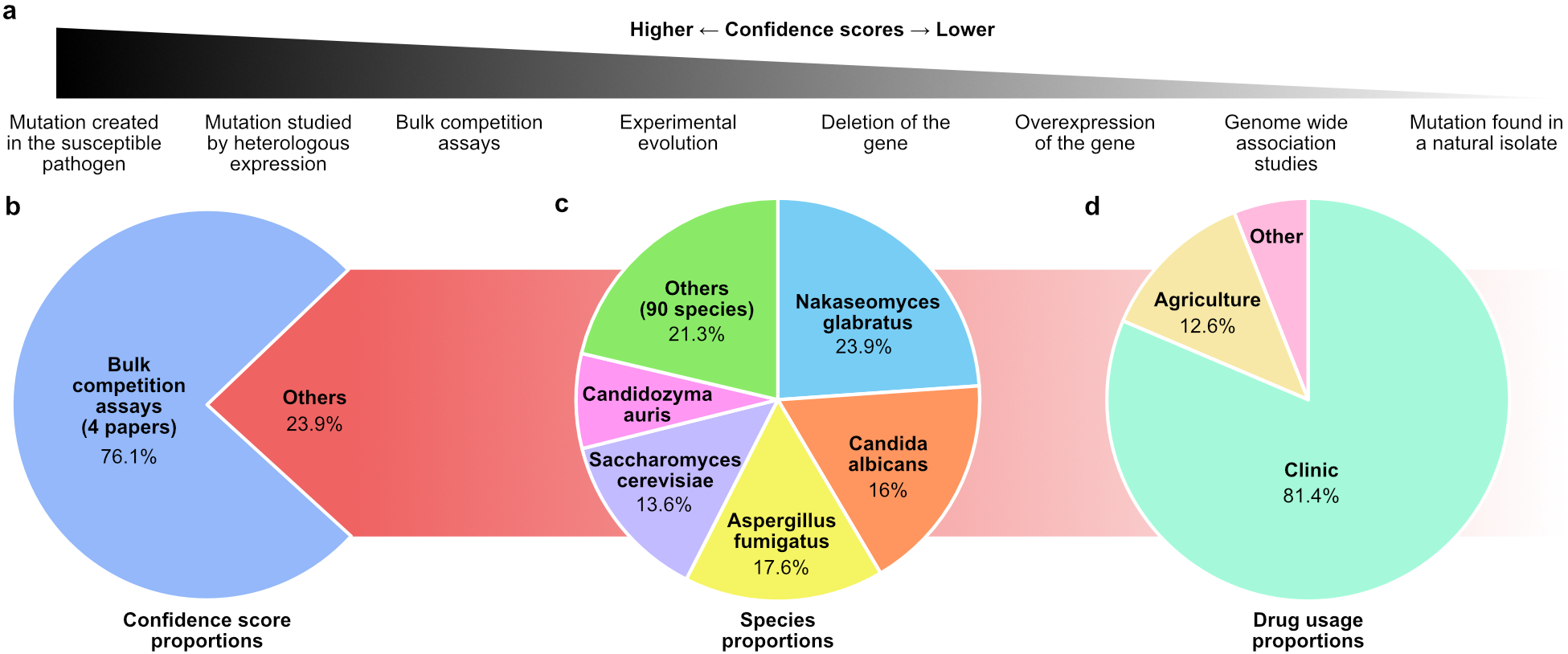
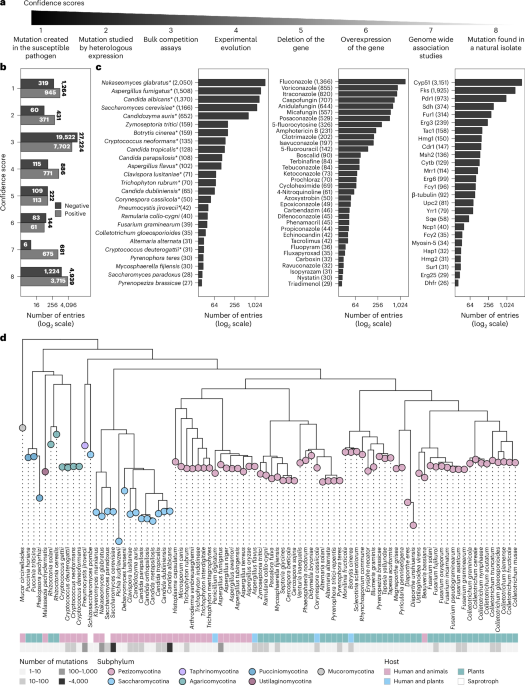
This one-year climb led to the FungAMR database3 containing 35,792 carefully curated entries across 208 drugs for 95 fungal species. To help researchers evaluate the strength of the association between a mutation and resistance, we assigned each mutation a confidence score based on the method of identification, ranging from the simple observation in a resistant strain to the experimental validation in a susceptible background (Figure 1a).
Once we finished the curation, we moved on to what I consider the fun part: analyzing the dataset. We took advantage of this newly curated database to establish the current state of knowledge in the field and to explore patterns of resistance across fungal species and antifungal classes. And like climbing a mountain, the higher the climb, the greater the view. We were therefore able to get an amazing view of the landscape of antifungal resistance.
Fungal AMR research is skewed toward a few species, genes and drugs
Despite reviewing over 500 articles, a significant portion of the data comes from a small number of studies that used high-throughput methods, such as bulk competition assays, to systematically map resistance mutations (Figure 1 b). This highlights how novel approaches can efficiently expand our understanding of resistance mutations across species and drug classes.
Also, although the number of entries is large, not all species and antifungals are well represented. Certain species tend to steal the spotlight, accounting for most of the entries in the database (Figure 1c). Much of this bias could come from the fact that AMR in human fungal pathogens is more frequently studied and reported than in plant pathogens. As a result, most of the antifungals covered are clinical drugs (Figure 1d) and the most studied proteins are their direct molecular targets.
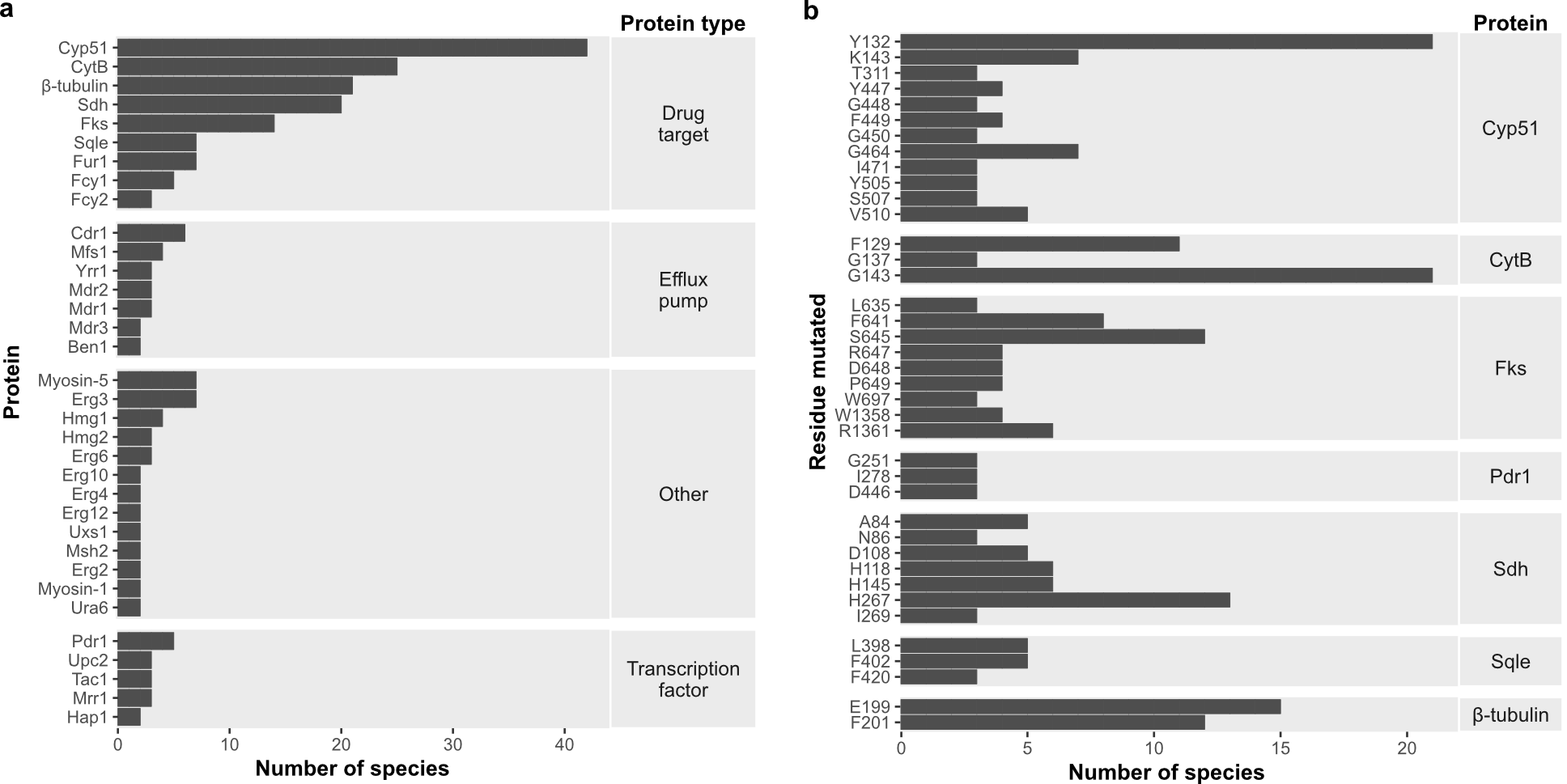
Some resistance mechanisms seem to be universal across species
Since FungAMR includes a large diversity of fungal species, we were able to compare resistance mutations across them. We found that many mutated genes (Figure 2a), and even specific resistance mutations within genes (Figure 2b), are shared between fungi that diverged hundreds of millions of years ago. This suggests that the ways resistance can be acquired may be limited and highlights the existence of potentially universal resistance mutations. Considering the biases in which species have been studied, this convergence has the potential to support the transfer of knowledge from well-studied fungi to less-studied pathogenic species.
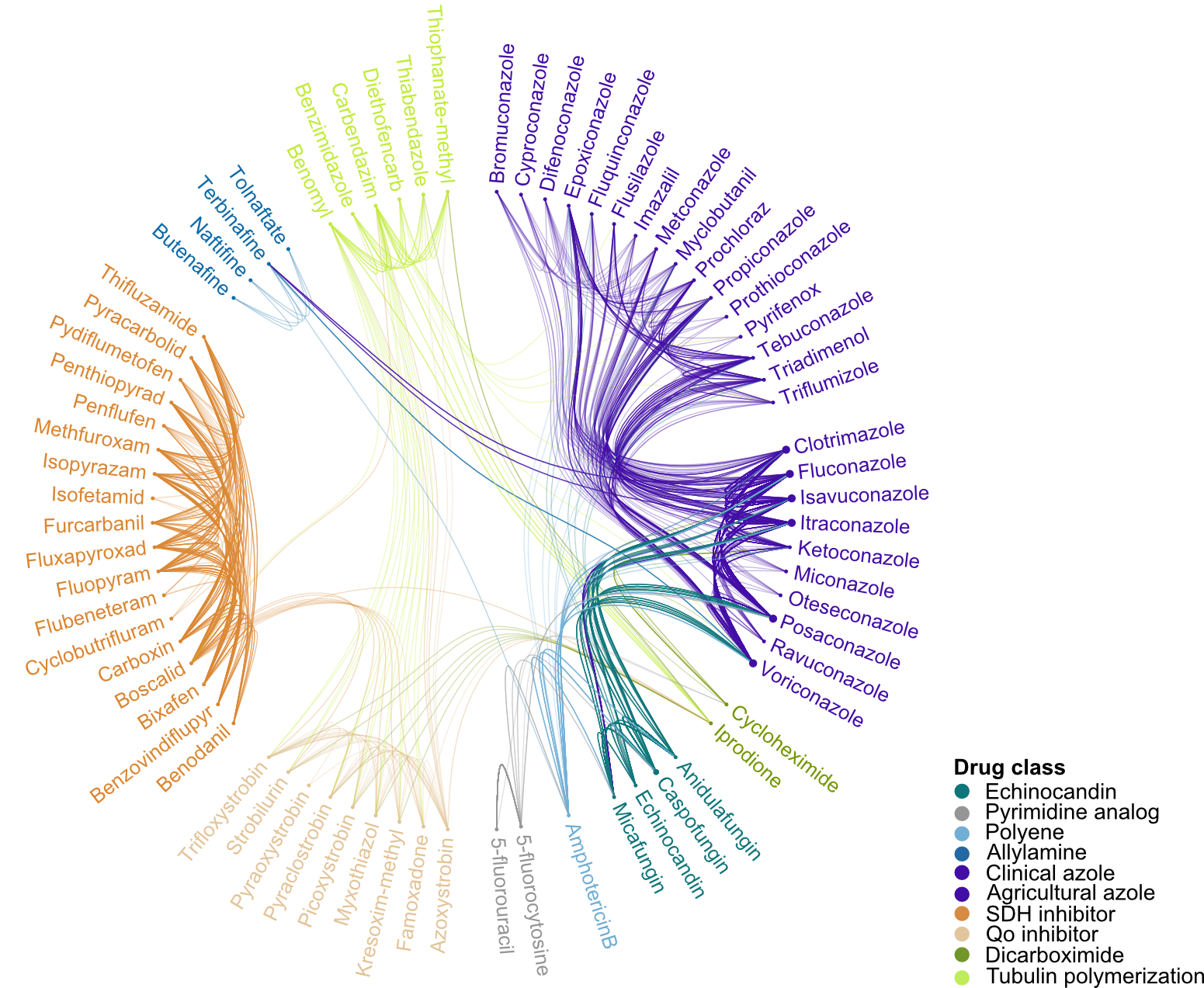
Cross-resistance between and within classes of antifungals is common
One major question in the field of AMR is whether mutations can confer cross-resistance (i.e. resistance to multiple drugs). With many mutations in the FungAMR database tested against more than one antifungal, we were able to assess the extent of this phenomenon. As expected, mutations that provide resistance to one drug often confer resistance to others within the same class. For example, we found widespread cross-resistance among azole drugs used in both clinical and agricultural settings. More notably, we also identified mutations that appear to confer resistance to more than one drug class, such as between azoles and echinocandins, which target different fungal enzymes and are typically considered alternative treatment options.
ChroQueTas can be used to screen AMR in fungal genomes
To facilitate the analysis of AMR in fungal genomes from the FungAMR data, we collaborated with Narciso M. Quijada and students from the University of Salamanca. They developed ChroQueTas, a user-friendly bioinformatic tool that can be used to detect AMR mutations in fungal genomes.
Collaboration with CARD and the community
FungAMR is a new resource for researchers studying antifungal resistance, offering a foundation for both basic and applied research. While not intended as a clinical decision-making tool, the dataset can support future efforts toward this goal.
To ease the use of database and its outreach to the scientific community, Andrew McArthur and its team from McMaster University kindly accepted to incorporate FungAMR as a web-searchable interface (FungAMR Mutation Data) within the Comprehensive Antibiotic Resistance Database (CARD)4.
We are planning to update FungAMR as new reports of fungal AMR are released. We encourage everybody to help us by filling up and sending us the same curation sheet the curators team used (Curation sheet). Any inquiries can be sent to the FungAMR e-mail address (fungamr.db@gmail.com).
References
1- Fisher, M. C. et al. A one health roadmap towards understanding and mitigating emerging Fungal Antimicrobial Resistance: fAMR. npj Antimicrob Resist 2, 1–10 (2024).
2-Nash, A. et al. MARDy: Mycology Antifungal Resistance Database. Bioinformatics 34, 3233–3234 (2018).
3- Bédard, C. et al. FungAMR: a comprehensive database for investigating fungal mutations associated with antimicrobial resistance. Nat. Microbiol. (2025).
4- Alcock, B. P. et al. CARD 2023: expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 51, D690–D699 (2023).
Poster image : Generated by Microsoft Designer AI (2025).
Follow the Topic
-
Nature Microbiology

An online-only monthly journal interested in all aspects of microorganisms, be it their evolution, physiology and cell biology; their interactions with each other, with a host or with an environment; or their societal significance.
Related Collections
With Collections, you can get published faster and increase your visibility.
The Clinical Microbiome
Publishing Model: Hybrid
Deadline: Mar 11, 2026
Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in