Zoonotic relevance of multidrug-resistant bacteria in parrots with respiratory illness
Published in Microbiology
Pet birds, the third most frequent companion animal after dogs and cats, are considered intimate pets of people and play an essential part in their lives. The majority of caged birds belong to two orders: Passeriformes, which includes canaries and finches, and Psittaciformes, which comprises parrots, parakeets, and lovebirds. Parrots are among the most valuable bird species worldwide. While parrots become infected with several pathogens of public health relevance, these infections are largely ignored compared to zoonoses of other companion animals. Many zoonotic pathogens are frequently isolated in passerines and parrots, including Enterobacter spp., Klebsiella spp., Escherichia coli, Serratia spp., Salmonella spp., Yersinia spp., and Staphylococcus aureus. Among Enterobacterales, pathogenic E. coli can induce both intestinal and extra-intestinal infections. The presence of E. coli strains in the guts of psittacine birds is a cause for concern owing to their risk of sepsis and mortality, as well as the possibility of transmission to human contacts. E. coli is responsible for nosocomial infections in humans, including catheter-related urinary tract infections and ventilator-associated pneumonia (VAP). K. pneumoniae could be detected in feces of parrots and passerines, but this pathogen is frequently regarded as a respiratory pathogen, especially in immunocompromised and stressed birds. In humans, K. pneumoniae is an opportunistic pathogen, causing both community-acquired and nosocomial infections, as well as UTIs, pneumonia, meningitis, sepsis, and pyogenic liver abscess. Among psittacine species, Salmonella Typhimurium is a commonly isolated Salmonella serotype, with outbreaks having been reported among birds. The pathogenicity of Salmonella is attributed to an array of virulence genes that are associated with clinical manifestations of Salmonella infection. P. mirabilis has been observed in pet birds, and it is the most widespread Proteus species associated with nosocomial infections and multidrug resistance. Furthermore, P. mirabilis causes food poisoning and extra-intestinal infections in people, mainly UTIs and others such as skin, wound infections, bloodstream infection, meningeoencephalitis, and osteomyelitis. Regarding Gram-positive bacteria, S. aureus is an opportunistic pathogen that induces omphalitis, bumblefoot, infected hocks, and stifle joints in birds. This bacterium lives on the skin and mucosa of healthy people; however, it is responsible for a wide range of life-threatening illnesses, such as food poisoning, skin disorders, and respiratory infections in humans.
A concern regarding the aforementioned pathogens is their resistance to various antimicrobials, complicating the treatment of these infections in birds, and potentially in humans if transmitted through animal contact. Antimicrobial resistance is a multifaceted phenomenon that poses a public health threat worldwide. Household pets and companion animals are identified as a significant source of multidrug-resistant zoonotic pathogens. Of particular concern in enterobacteria is resistance to cephalosporins, which is mediated by extended-spectrum beta-lactamase (ESBL) genes carried by transferable plasmids. Also, methicillin-resistant Staphylococcus aureus (MRSA) is a superbug pathogen in human and veterinary medicine, and multidrug resistance in MRSA is a main therapeutic challenge worldwide. Bacterial pathogens isolated from parrot cloaca are a common research focus. However, little is known regarding the nasal carriage of zoonotic pathogens in parrots suffering from respiratory illness, which is transmitted mainly by droplets and aerosols, posing a risk for human contacts. Thus, the main purpose of this study was to investigate the occurrence of multidrug-resistant bacteria with zoonotic potential in parrots suffering from respiratory illness.
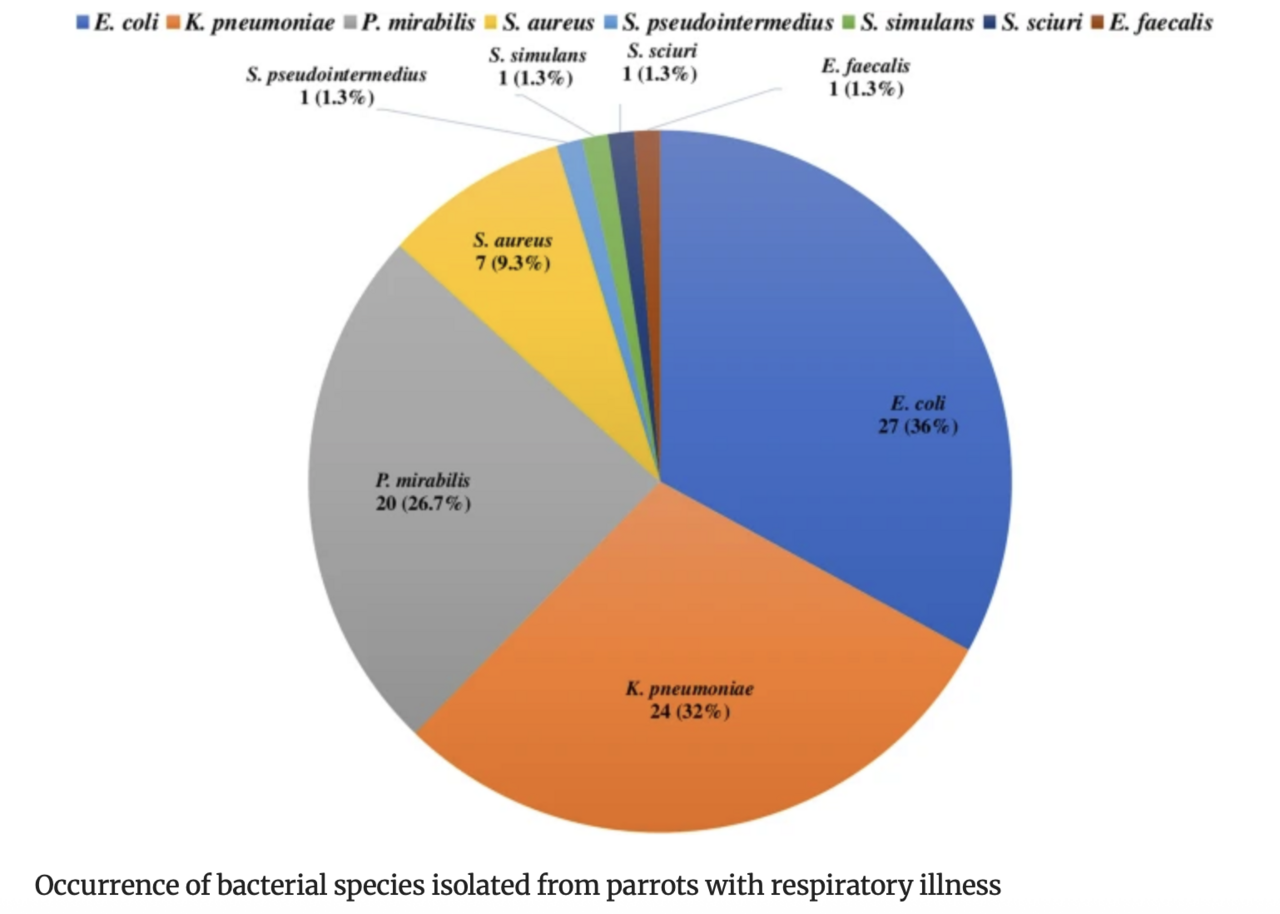
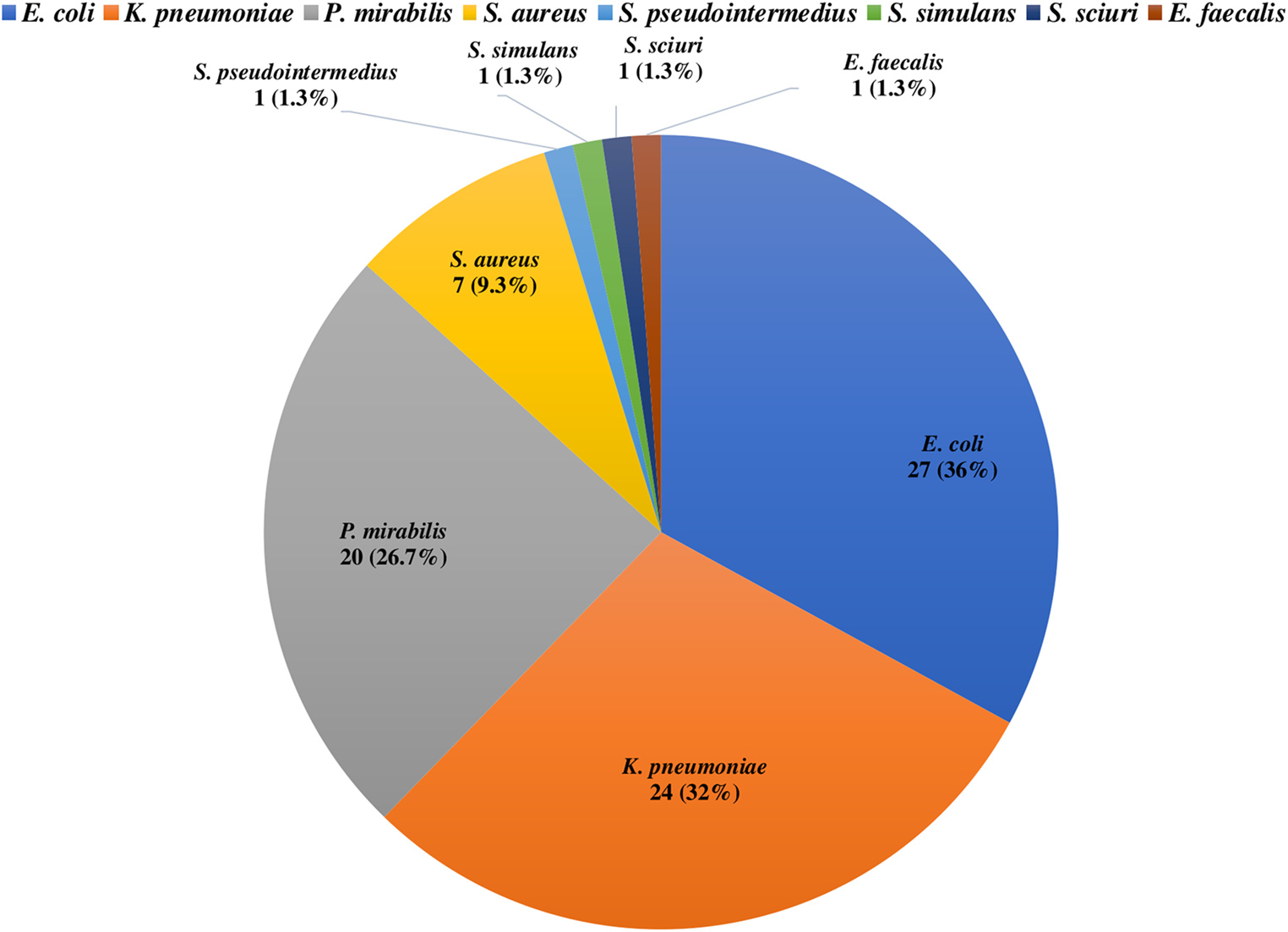
Nowadays, research attention is paid to the investigation of bacterial pathogens in the cloaca of parrots rather than the nasal niche, which is largely ignored. Therefore, this study aimed to investigate the nasal carriage of multidrug-resistant bacteria with zoonotic potential in parrots suffering from respiratory illness. Nasal swabs were collected from 75 sick parrots with respiratory illness, and they were subjected to microbiological isolation and identification, followed by antimicrobial susceptibility testing. Escherichia coli, Klebsiella pneumoniae, Proteus mirabilis, and Staphylococcus aureus were isolated with a prevalence rate of 36%, 32%, 26.7%, and 9.3%, respectively, while one isolate (1.3%) of Staphylococcus pseudointermedius, Staphylococcus simulans, Staphylococcus sciuri, and Enterococcus faecalis was identified. E. coli, K. pneumoniae, and P. mirabilis were investigated for ESBL genes, Staphylococcus species for the mecA gene, followed by SCCmec typing, and E. faecalis for the vanA and vanB genes. Regarding beta-lactamase-encoding genes, blaTEM(97.6%), blaSHV (48.8%), and blaCTX−M (39%) gene families were detected, while blaOXAwas not found. Sequencing of blaCTX−M in one strain of E. coli, K. pneumoniae, and P. mirabilis revealed blaCTX−M−15. The mecA was determined in three S. aureus and one S. sciuristrain, and the SCCmec typing of three MRSA isolates yielded type V, whereas type I in S. sciuri. Only the vanA gene was recognized in the E. faecalis strain. Moreover, 67.1% of bacterial isolates exhibited multidrug resistance. These findings highlight the potential role of parrots in the transmission of multidrug-resistant zoonotic bacteria, which may pose a threat to human contacts.
In conclusion, the recent study highlights the nasal carriage of multidrug-resistant zoonotic bacteria among parrots with respiratory illness, indicating that parrots may be a source of antimicrobial-resistant bacteria to pet owners and emphasizing the importance of enhanced global surveillance for pet bird-related antimicrobial resistance. A limitation of the current study is that we did not perform whole genome sequencing (WGS) due to a lack of resources. WGS provides a broader spectrum of genomic information, enabling a more comprehensive characterization of antimicrobial-resistant bacteria.
Follow the Topic
-
Veterinary Research Communications

This journal publishes fully refereed research articles and topical reviews on all aspects of the veterinary sciences. Interdisciplinary articles are particularly encouraged, as are well argued reviews, even if they are somewhat controversial.

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in