The Evolutionary Game of Alpha-helical Protein Residues
Published in Cancer, Chemistry, and Ecology & Evolution
Check our full behind the paper story here: https://go.nature.com/3KnbhdB
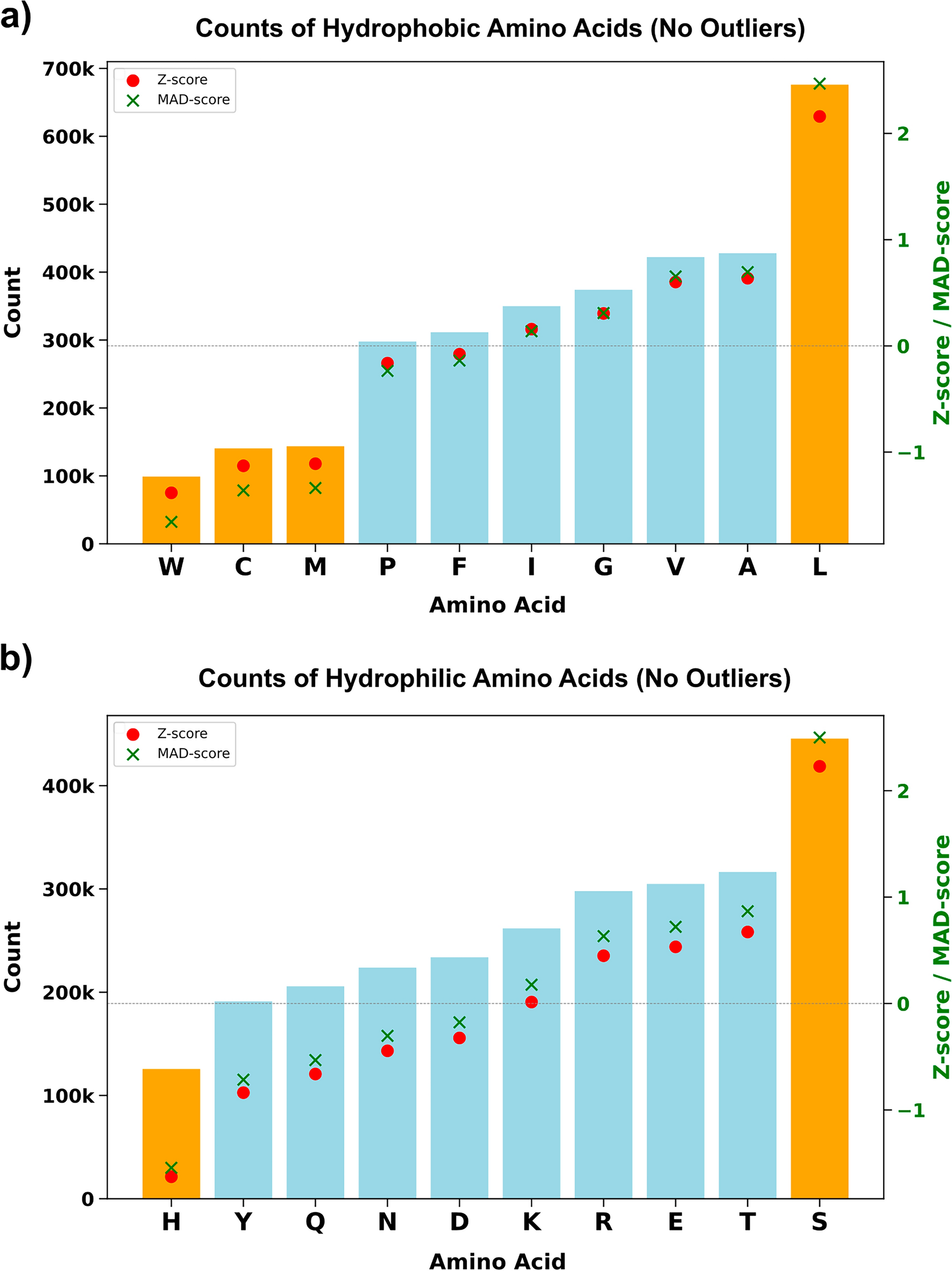
Our interest in membrane proteins began with a practical challenge: how do you study proteins that are so hard to solubilize and crystallize? Hydrophobic α-helices hide in membranes, making structural work notoriously difficult. In our early work on the QTY code, we asked: can we swap hydrophobic residues (L, I, V, F) with polar ones (Q, T, Y) in neurological transporters to make them soluble without disrupting their helices?
Surprisingly, the answer was yes (Zhang et al. PNAS 2018; Karagöl et al. Pharm Res 2024; Karagöl et al. PLOS One 2024a; Johnsson et al. 2025). The substitutions preserved fold and even function, showing that evolution tolerates more flexibility than textbooks suggest. This raised a deeper question: could evolution itself exploit similar hydrophobic–polar swaps to diversify function, adapt to environments, or buffer harmful mutations?
Follow the Topic
-
Journal of Molecular Evolution

This journal covers experimental, computational, and theoretical work aimed at deciphering features of molecular evolution and the processes bearing on these features.



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in