A manager of reactions
Published in Chemistry

How many reactions can be run at once? With the advent of data-driven models and a thirst to rapidly discover high impact reactivity, this question drove the experimental infrastructure of our lab. A well organized scientist may diligently track many experiments, but dedicated software is needed to maximize throughput and maintain results in a standardized, computer-readable format. We realized there was no trivial or accessible way to design and manage plated reaction arrays and decided to develop phactor™, named in part due to our association with the University of Michigan's College of Pharmacy. With standardized hardware existing from high throughput screening techniques used in biological assays, phactor™ works with size 24, 96, 384, and 1,536 wellplates.
phactor™ was designed to minimize the number of clicks needed to take a chemist from experiment idea to reaction results. With feedback from over one hundred chemists who have used the software, the user experience allows even novice scientists to create and execute robust yet flexible reaction arrays. The optimized workflow is split into six stages: settings, factors, chemicals, grid, analysis, and report. We realized much of the design process can be automated in the software.
On the settings and factors stage, the user names their experiment, dictates the throughput and other experimental metadata such as temperature and stir rate, and inputs the experimental factors that will be screened in the multiplexed array, such as the number of catalysts and ligands that will be cross-tested in the reaction plate. With this information, phactor™ can automatically distribute the well locations of each reagent - ensuring a full combination of all experimental factors. The user then inputs reagents on the chemicals stage until all the expected factors for the experiment are satisfied. Reagents can be input manually with associated molecular weights and names, or through a variety of interfaces including external database connectivity or even artificial intelligence based GPT widgets. When all the reagents have been added, an experimental design is automatically generated. This is viewed on the grid stage, where single or bulk edits can be made to the experiment via an interactive grid representing the wellplate. On this stage, the chemist can download the step-by-step recipe to make stock solutions to be distributed into the wellplate or interface with liquid-handling robots to automate the dosing process. Once the reactions are complete, quick analytics can be performed on the analysis stage and experimental inputs and outputs can be downloaded in a machine readable and standardized format on the report stage.

At this point, phactor™ has been used extensively in our lab. We have a strong interest in developing various amine-acid couplings and phactor™ has allowed us to test many conditions using these abundant building blocks that exists in our inventory. To date, phactor™ has been used to discover two amine-acid esterification reactions, three amine-acid C–C couplings, and a variety of optimized conditions for amide couplings. For many of these experimental campaigns, phactor™ was instrumental in initial reaction discovery, reagent optimization, and the expansion of the reactivity's substrate scope. In our ultrahigh throughput esterification reaction, phactor™ demonstrated the substrate versatility of the method in a size 1,536 wellplate.
phactor™ has also been used in optimizing steps of total syntheses and in direct-to-biology assay development. In fact, phactor™ was used to discover a novel and competitive SARS-CoV-2 Main Protease inhibitor through an ultrahigh throughput direct-to-biology campaign. After an initial 24-well exploratory experiment testing the viability of the chemistry and biology, an inhibitor library was synthesized using amide chemistry on a 1,536 wellplate. With our collaborators in the Life Sciences Institute, each reaction in the wellplate was sampled and tested for inhibition against the target protein. phactor™ tied the chemical and biological results together, and the best hits were scaled up and isolated.
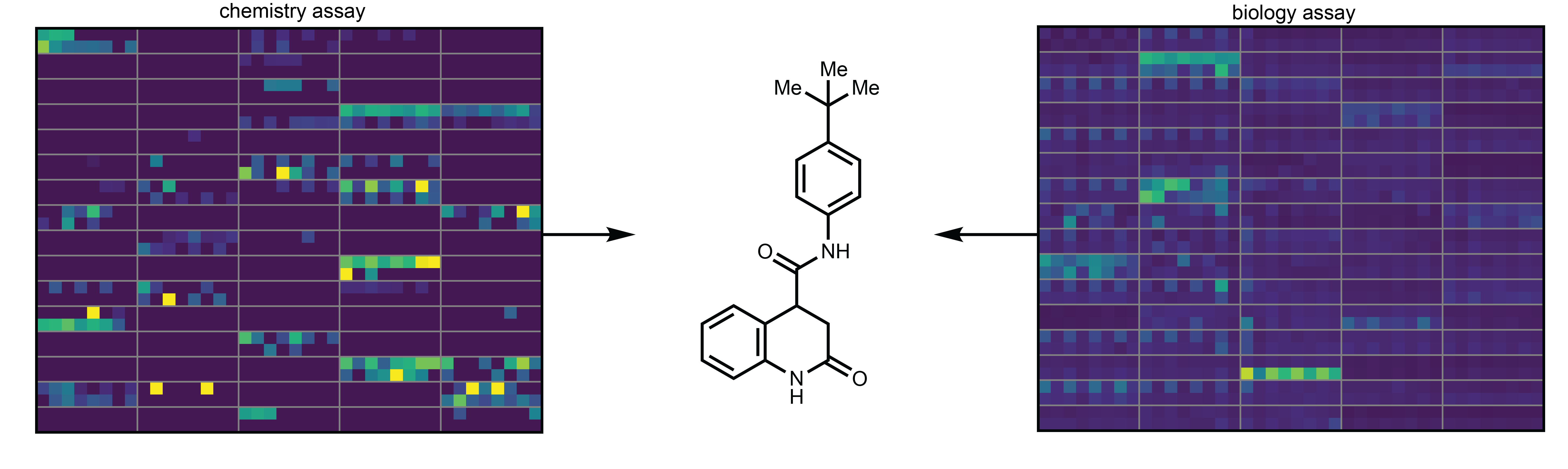
So how many reactions can be ran at once? It's hard to exactly know, but phactor can help answer the question by maximizing our throughput and letting chemists focus on the chemistry. Link to the article here: https://www.nature.com/articles/s41467-023-39531-0
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in