Adaptive alleles stick together
Published in Ecology & Evolution

The ecological drivers of local adaptation in plants are often easy to understand: evolve salt-tolerance to grown next to the sea, change flower colour to attract different pollinators, and so on. However, how that is achieved at the genetic level remains, most of the times, a mystery.
Wild sunflowers make for a particularly good model system to study the genetic basis of adaptation. The wild sunflower family is large and diverse, and includes dozens of species that grow across North America; they are adapted to a wide range of environments, including rather extreme ones like deserts, salt marshes and serpentine soils, and well-characterized ecotypes are known in many of these species. Additionally, thanks to their connection to cultivated sunflower, there are more genetic and genomic resources available for (wild) sunflowers than for most other ecological models. Yes, they have very repetitive, 3+ Gbp genomes, but nobody’s perfect1.
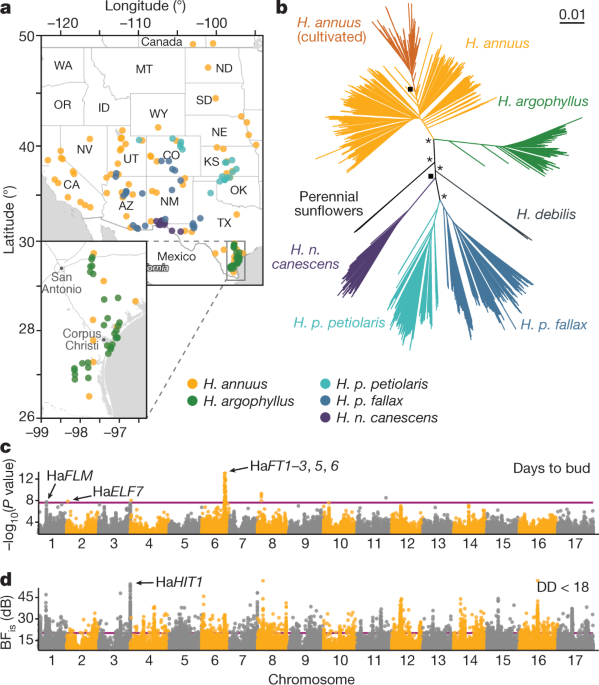
To understand how wild sunflowers managed to adapt so well to difficult environments, we phenotyped in a common garden and resequenced 1500+ plants. To our surprise, we found not only individual genes, but also massive regions of the genome (up to 100+ Mbp) tightly linked to adaptive traits, and to environmental variables (climate and soil composition). Many of these regions are huge, segregating inversions; by suppressing recombination, they keep together combinations of adaptive alleles that would otherwise be recombined apart when non-adaptive alleles are introduce through gene flow from nearby, non-adapted populations. These non-recombining haplotypes (which we called haploblocks, to account for the fact that a few of them do not appear to be inversions) explain several well-known cases of local adaptation in wild sunflowers.


How did we get here?
This is a really neat story, and it closely aligns with evolutionary theory predicting a major role of inversions and other recombination modifiers in generating and maintaining ecotypic diversity. All of this is (hopefully) clearly described in our paper2. What might not transpire from the paper is the ridiculous amount of work, from many talented people, that went into creating the datasets behind those snazzy plots.
The first step was getting a comprehensive collection of seeds for the three sunflower species we wanted to focus on. While the USDA and research labs (including ours) hold collections for several wild sunflower species, some are decades-old, have been propagated in cultivated fields, and/or have spotty collection records. In the summer of 2015, an adventurous postdoc in our lab (Dylan Burge) started an epic four-months field trip across North America (with a shorter additional leg in 2016). Driving almost 50,000 kilometers, through deserts and snow storms, he brought back a collection of seeds from several thousand sunflowers which became the foundation of our study. He also mailed half a ton of soil sampled from collection sites across the U.S.



Once the seeds arrived in Vancouver (where our lab is based), another adventure started. Wild sunflowers are unruly, bushy things, that can grow up to 4 meter tall and more than 2 meters wide – a field with 1500 of them quickly becomes an impenetrable jungle. Throughout eight months, we germinated, transplanted, crossed and phenotyped daily all the plants, with the help of many colleagues and dedicated students in the lab (all of whom we are extremely grateful to).


Surplus sunflowers (including some cultivars that we used as border plants, like the teddy bear sunflower in the foreground) were regularly donated to lucky passersby - this made us pretty popular within the UBC campus community.


Even after the plants were dead, we had enough digital images to keep us busy for another two years, extracting phenotypic data (which included fun activities like counting trichomes and ligules, measuring flowers and seeds, etc.). Some of this work more than paid off - measures of flowering time or seed size resulted in the spectacular GWA peaks and plateaus that are feature in our paper. Others, like trichome density or leaf shape, were a complete bust (or, as one would say in a paper, "highly polygenic"). At the same time, countless hours and energies were spent extracting DNA, preparing sequencing libraries from all 1500 plants, and making sense of the 14.5 Tbp of sequence data we got out of them – programs designed for relatively homogenous human sequence data faltered when confronted with highly diverse sunflower species, and even mundane, well-established steps in our genotyping pipeline required extensive troubleshooting and optimization.

Where do we go from here?
We now know a lot more about these haploblocks, and more in general about the genetic/genomic basis of adaptation in wild sunflowers, than we knew five years ago. However, with great knowledge comes a great number of new questions; a few are listed below, in no particular order.
- What is in these haploblocks? Despite having a ton of sequence data, we still can’t be sure about what exactly is in these regions. Haplotypes at an haploblock region can be very divergent (they share on average ~95% similarity, lower than that between humans and orangutan), and we have to rely on SNP information based on the reference assembly for cultivated sunflower to try to understand what is in them. There could be dozens of new genes in each haploblock, which could include those responsible for the adaptive variation we observed. Obtaining highly contiguous assemblies for several wild sunflowers is likely going to be the only way to figure that out.
- Where do they come from? In a few cases, we can see that one of the haplotypes at an haploblock region derived from introgression from a different sunflower species. For most haploblocks, however, where the different haplotypes come from or how they were generated is not clear; introgression seems the most plausible explanation in these cases as well, but we haven’t found the donor species yet. One favorite pet theory is that haploblocks are remnants of extinct sunflower species that were adapted to those habitats – a new sunflower species came in, hybridized with them, “stole” their adaptive modules (the haploblocks), and replaced them.
- Is there more of them? Most certainly. Our re-sequencing approach could only identify highly divergent haploblocks larger than 1 Mbp. Smaller and/or less divergent structural variants affecting adaptive traits could be much more numerous.
- Could you use them for sunflower improvement? That was one of our first thought – if you want to grow a field of sunflowers in the desert, you pop in the corresponding adaptive module (aka haploblock), and voilá. Even when introducing individual genes from wild relatives things are of course not that easy, but haploblocks come with additional challenges. It is true that the “adaptive” genes within the haploblocks could help cultivated sunflower survive in harsher environments. However, haploblocks are huge, and can contain thousands of other genes – some of them could very well have an effect that is detrimental in a cultivated setting, or that negatively affects yield. If the introduced haploblock region cannot recombine with its cultivated counterpart, this linkage drag would be impossible to break. Paradoxically, knowledge of the existence of these haploblock could instead be more useful to avoid them when using wild relatives in sunflower breeding programs. Indeed, some haploblocks are already segregating within the cultivated sunflower germplasm.
- Are they a sunflower thing? That seems very unlikely – cases of individual large, old structural variants linked to adaptation have been reported before (for example in cod and salmon3,4). Current sequencing technologies make it hard to detect such large haplotype/structural variants, unless they are extremely divergent (as is the case for sunflower haploblocks) and you have a thorough representation of a species genetic diversity; as long reads sequencing technologies and techniques to map long-range chromatin interactions become more commonplace, we’ll likely start to see more and more evidence of a widespread role of structural variants in adaptation.
In the end, all the hard work was of course well worth it - the combination of genetic, phenotypic and environmental data opened up new lines of research that will keep us busy for years to come; it is quite clear that we only started to understand the role of structural variants in adaptation. There are also a lot of cool, non-haploblocks-related results that came out of these datasets, and that we are still exploring (individual adaptive genes, patterns of divergence and selection...). For now, however, we are also just happy that there are no more trichomes to be counted.
- Billy Wilder, Some like it hot (1959), United Artists.
- Todesco M., Owens G.L., Bercovich N. et al. Massive haplotypes underlie ecotypic differentiation in sunflowers (2020), Nature, https://doi.org/10.1038/s41586-020-2467-6.
- Pearse D.E., Barson N.J., Nome T. et al. Sex-dependent dominance maintains migration supergene in rainbow trout. Nat Ecol Evol 3, 1731–1742 (2019).
- Kirubakaran T.G., Grove H., Kent M.P., Two adjacent inversions maintain genomic differentiation between migratory and stationary ecotypes of Atlantic cods. Mol Ecol 25, 2130–2143 (2016).
Follow the Topic
-
Nature

A weekly international journal publishing the finest peer-reviewed research in all fields of science and technology on the basis of its originality, importance, interdisciplinary interest, timeliness, accessibility, elegance and surprising conclusions.





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in
a really entertaining story (and fascinating results) Natalia! thank you!
It's a so wonderful post and contains so many beautiful photos!~
There probably occurs a misspell: he prairie sunflower, The prairie sunflower.
Thanks Gao! Also for spotting the typo - the "T" got lost while copy-pasting the figure legend.