Altered RNA export by SF3B1 mutants confers sensitivity to nuclear export inhibition
Published in Cancer, Cell & Molecular Biology, and Genetics & Genomics
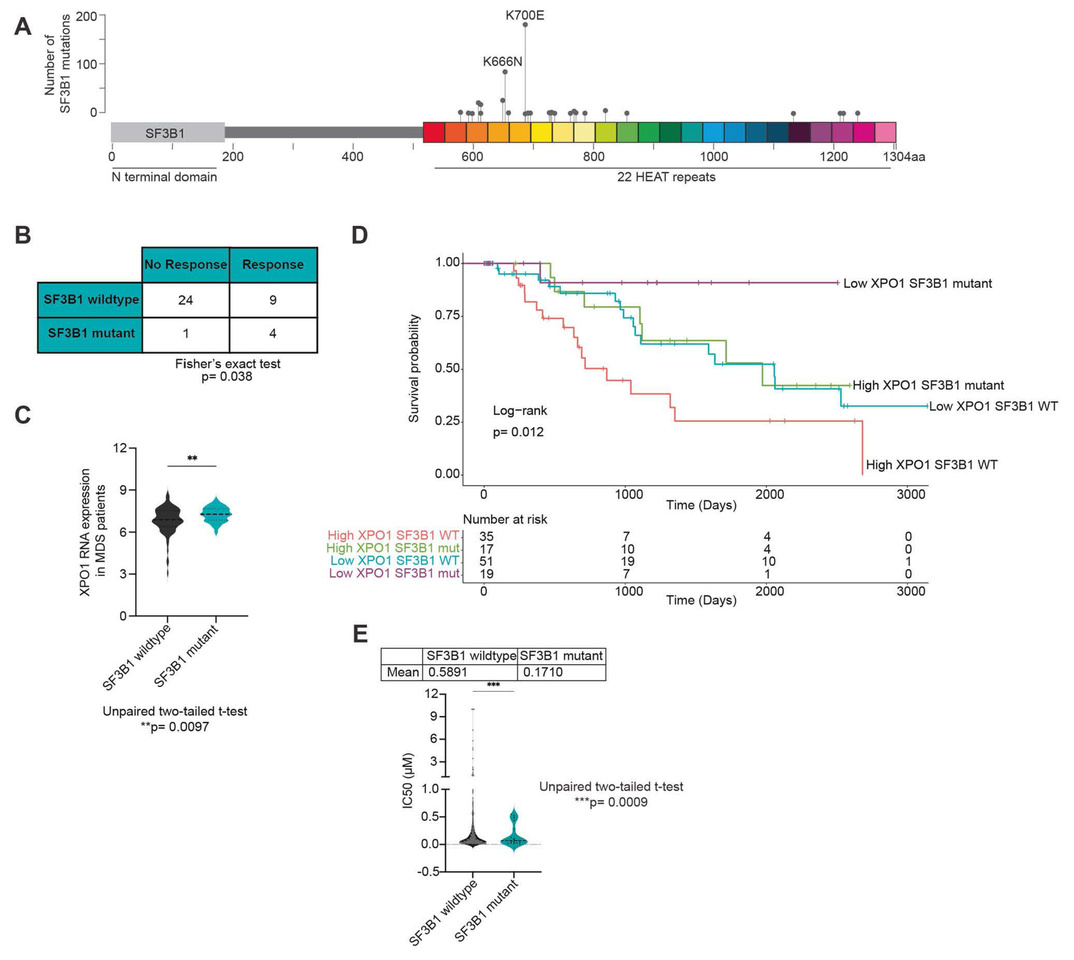
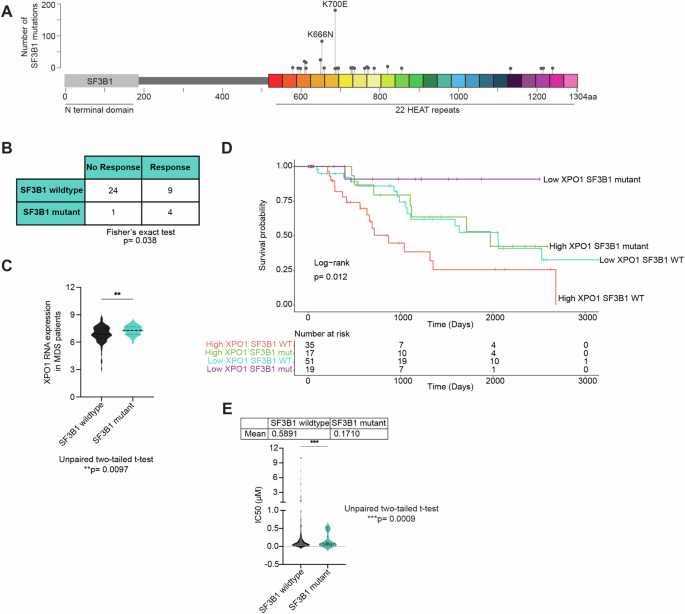
SF3B1 mutations, the most frequently observed splicing factor gene mutations seen in various cancers, have long posed a challenge for targeted therapy development due to the requirement for splicing in normal biology. Recent clinical trials for drugs targeting nuclear export have highlighted that patients with high-risk myelodysplastic neoplasms (MDS) harboring SF3B1 mutations showed enhanced responsiveness to XPO1 inhibitors, such as selinexor and eltanexor. XPO1, or Exportin-1, is crucial for the transport of proteins and RNA species out of the nucleus, which is integral to maintaining cellular homeostasis and gene expression.
Our research paper explores this phenomenon in further detail. We hypothesized that SF3B1 mutations alter cellular responses to XPO1 inhibition, potentially due to changes in splicing via nuclear export of small nuclear RNAs that are involved in spliceosomal assembly. Our RNA sequencing analysis revealed that after XPO1 inhibition, SF3B1-mutant cells exhibited increased nuclear retention of RNA transcripts and a shift towards 3'SS and SE alternative splicing, particularly affecting genes involved in apoptosis. Furthermore, snRNAs were retained within the nucleus of SF3B1 mutant cells after XPO1 inhibition.
To extend these findings into therapeutic strategies, we performed a forward genetic screen to identify drug combinations that might enhance the efficacy of XPO1 inhibitors. The results pointed to BCL2 and BCLXL, anti-apoptotic proteins, as promising targets. Subsequent in vivo experiments with Sf3b1K700E mutant mice demonstrated that combining eltanexor with venetoclax (a BCL2 inhibitor) selectively targeted SF3B1 mutant cells with minimal off-target toxicity.
Combination strategies such as the one proposed in our paper could have promising results for improving treatment outcomes in high-risk MDS. These need to be tested further in clinical trials before they could be recommended for treatment in patients.
Follow the Topic
-
Leukemia

This journal publishes high quality, peer reviewed research that covers all aspects of the research and treatment of leukemia and allied diseases. Topics of interest include oncogenes, growth factors, stem cells, leukemia genomics, cell cycle, signal transduction and molecular targets for therapy.


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in