An evolving landscape for development and studies of antibodies in allergy at the molecular level
Published in Immunology
Antibodies are wonderful partners in life that protect us from threats found in the environment, like infectious agents and toxins. These proteins, also known as immunoglobulins, are made up of two types of protein chains, the heavy and the light chain, that combine to enable recognition of a vast diversity of targets, such as bacterial proteins. Other parts of antibodies directly interact with cells and other molecules of the immune system to destroy the danger. As they are highly potent, an elaborate system of cells and molecules controls the development of antibodies in order to promote the emergence of antibody responses to treats and to prevent the emergence of antibodies destructive to our health. Such systems not only define the development of antibodies, but also which type of antibody that is to be produced. Humans produce five antibody classes, IgM, IgD, IgG, IgA, and IgE, and in addition different variants (sub-classes) of IgG and IgA. These different antibody classes and sub-classes have different roles in immunity and interact with other parts of the immune system. For instance, IgA is typically active on mucosal surfaces where they act to protect against invasion, and IgE may have a role in protection against parasites. Sometimes, these control mechanisms fail, resulting in conditions caused by the presence of antibodies that carry out inappropriate reactions. This may manifest itself for instance as autoimmunity, conditions during which antibodies attack your own molecules and tissues, or allergy. Altogether, there are multiple reasons to study antibodies, their complexity and development and their roles in health and disease. The vast diversity of antibodies imposes particular challenges for such studies, challenges that must be addressed.
Allergic disease typically develops when antibodies of a specific class, IgE, are inappropriately formed, commonly in response to encounter with otherwise harmless agents like pollen and foods, the so-called allergens. IgE antibodies are in comparison to other classes of antibodies like IgA, IgG, and IgM present in minute quantities in blood. Yet they can induce the very challenging, sometimes even life-threatening, symptoms that substantially affect the quality of life of allergic subjects. In any case, the rarity of IgEs also represents a substantial challenge as they become difficult to investigate. Indeed, IgE was the last class of human antibodies to be discovered. The discovery of IgE during the second half of the 1960ies, however, enabled rapid development of tools and novel technologies that allowed for diagnosis and characterization of the allergic immune responses. Subsequent use of recombinant DNA technology allowed for detailed characterization of the many allergenic proteins that are recognized by the allergic subjects’ IgE. The low abundance of IgE in clinical samples and the extreme rarity of cells producing this type of antibody still severely hampered the generation of reagents and resources that would allow us to develop a more complete understanding of IgEs in disease and their interactions with the allergens. A recombinant antibody technology, an approach awarded the Nobel Prize in Chemistry in 2018, and human hybridoma technology, an extension of another Nobel Prize-winning scientific discovery, has nevertheless allowed us to in part address details of the biology of IgE in greater detail. Importantly, the emergence of novel technologies over time offers opportunities to allow us to establish an enhanced understand antibody responses, including those that are central to the allergic condition.
Further high throughput technology development, in particular next generation sequencing (NGS) and lately, single-cell sequencing, now allows us to assess the diversity of antibody responses, in allergy and beyond, in very great detail. Such methods and associated computational analysis platforms have come to play a substantial role in our efforts to define such antibody responses. In a recent study (Thörnqvist et al., 2026) we demonstrate an approach to exploit such advanced technologies in combination (Figure 1), to define antibodies of the IgE class directly from allergic subjects’ antibody-producing cells. In contrast to past studies, this was achieved without a preconception of the allergenic molecules involved in the allergic response of study subjects, in essence enabling identification of a wider range of IgEs. By employing single-cell sequencing we can indeed not only identify cells producing allergen-specific antibodies but also further develop reagents based on such antibodies to grow our understanding of allergy-causing immunity in humans. This opens up for a research field that in detail identifies and exploits allergic subjects’ immune responses to develop novel strategies in allergy diagnosis and, importantly, in vaccination against allergic disease, the only treatment with truly curative potential in allergy. 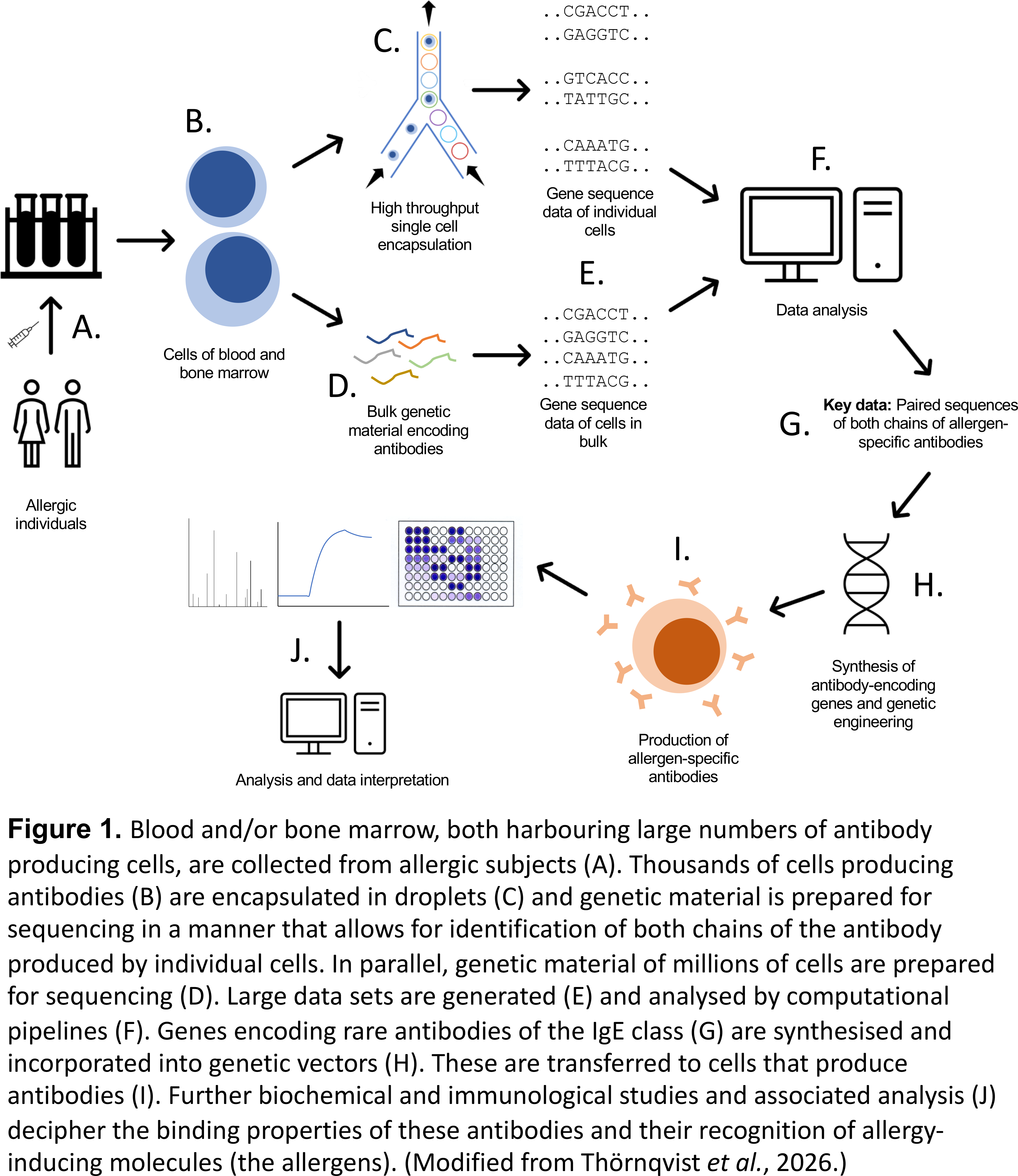
Altogether, this development is part of a long journey, enabled by a continuous technical development in the field of molecular immunology, gene sequencing, and bioinformatics/AI/ML (a few aspects of which are mentioned in Figure 2) that impact us all. Incorporation of novel tools and certainly also adaptation and evolution of already established technological platforms into IgE research are, despite challenges associated to their cost, critical for the future stepwise evolution of knowledge of the allergic condition.
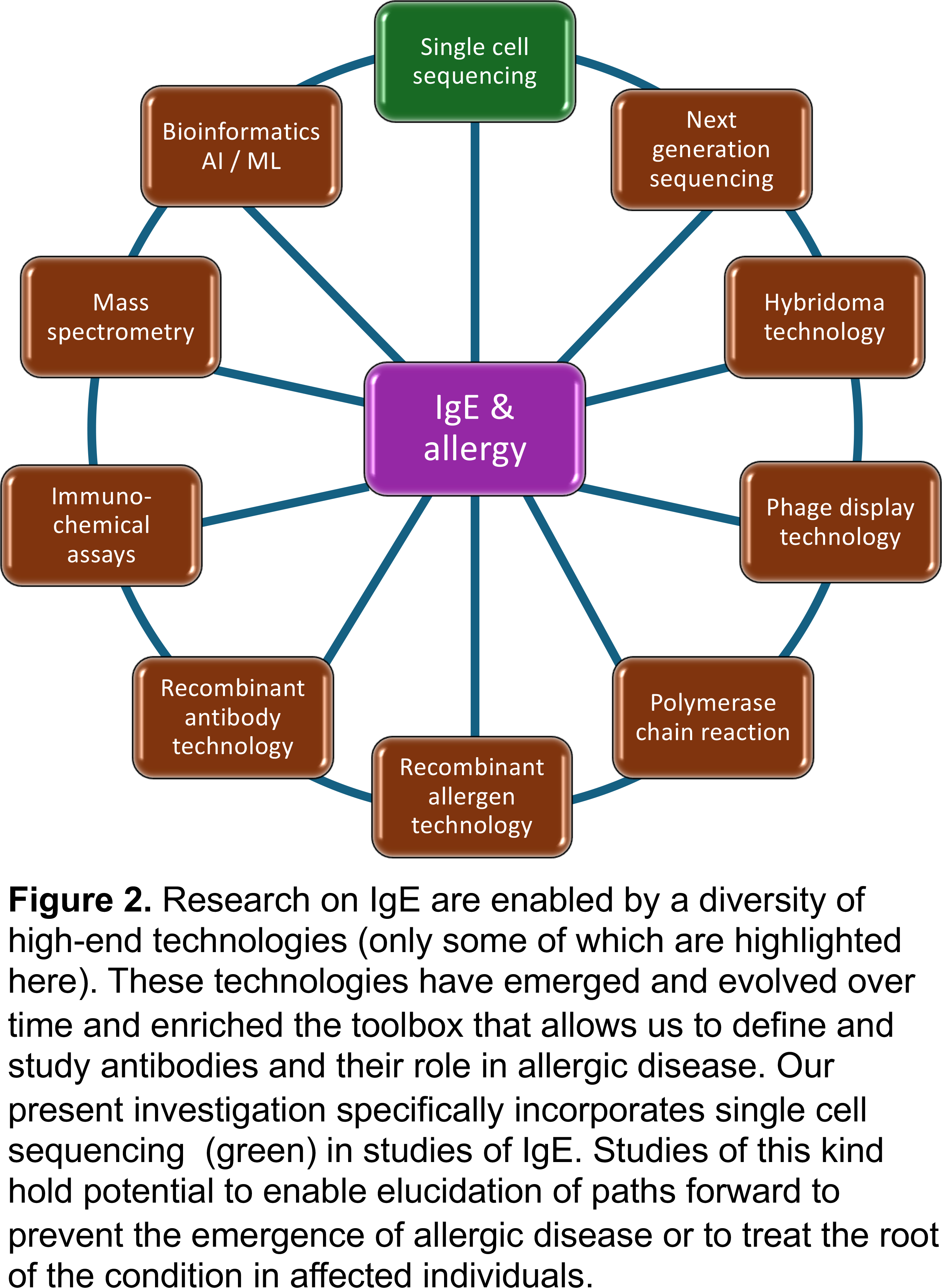
If you would like to know more, please use these links to access related reading on the different aspects and evolution of our IgE research pipeline:
- Essén M et al. (2025) Low nanomolar affinity to major grass pollen allergen Phl p 5 as achieved in an unmutated human antibody-lineage ancestor. Front Immunol 16, 1600778.
- Hoh RA et al. (2023) Clonal evolution and stereotyped sequences of human IgE lineages in aeroallergen-specific immunotherapy. J Allergy Clin Immunol 152, 214-229.
- Thörnqvist L et al. (2022) Linear epitope binding patterns of grass pollen-specific antibodies in allergy and in response to allergen-specific immunotherapy. Front Allergy 3, 859126.
- Mikus M et al. (2021) Allergome-wide peptide microarrays enable epitope deconvolution in allergen-specific immunotherapy. J Allergy Clin Immunol 147, 1077-1086.
- Glesner J et al. (2019) A human IgE antibody binding site on Der p 2 for the design of a recombinant allergen for immunotherapy. J Immunol 203, 2545-2556.
- Levin M et al. (2017) Antibody-encoding repertoires of bone marrow and peripheral blood-a focus on IgE. J Allergy Clin Immunol 139, 1026-1030.
- Levin M et al. (2016) Persistence and evolution of allergen-specific IgE repertoires during subcutaneous specific immunotherapy. J Allergy Clin Immunol 137, 1535-1544.
- Levin M et al. (2014) Multiple independent IgE epitopes on the highly allergenic grass pollen allergen Phl p 5. Clin Exp Allergy 44, 1409-1419.
- Levin M et al. (2014) Human IgE against the major allergen Bet v 1 - defining an epitope with limited cross-reactivity between different PR-10 family proteins. Clin Exp Allergy 44, 288-299.
- Levin M et al. (2013) Phl p 1-specific human monoclonal IgE and design of a hypoallergenic group 1 grass pollen allergen fragment. J Immunol 191, 551-560.
- Persson H et al. (2007) Delineating the specificity of an IgE-encoding transcriptome. J Allergy Clin Immunol 120, 1186-1192.
- Andréasson U et al. (2006) The human IgE-encoding transcriptome to assess antibody repertoires and repertoire evolution. J Mol Biol 362, 212-227.
Follow the Topic
-
Communications Biology

An open access journal from Nature Portfolio publishing high-quality research, reviews and commentary in all areas of the biological sciences, representing significant advances and bringing new biological insight to a specialized area of research.
Related Collections
With Collections, you can get published faster and increase your visibility.
Lipids in Cell Biology
Publishing Model: Open Access
Deadline: Mar 03, 2026
Forces in Cell Biology
Publishing Model: Open Access
Deadline: Apr 30, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in