Angiogenic and immune predictors of neoadjuvant axitinib response in renal cell carcinoma with venous tumour thrombus
Published in Cancer, Computational Sciences, and Biomedical Research

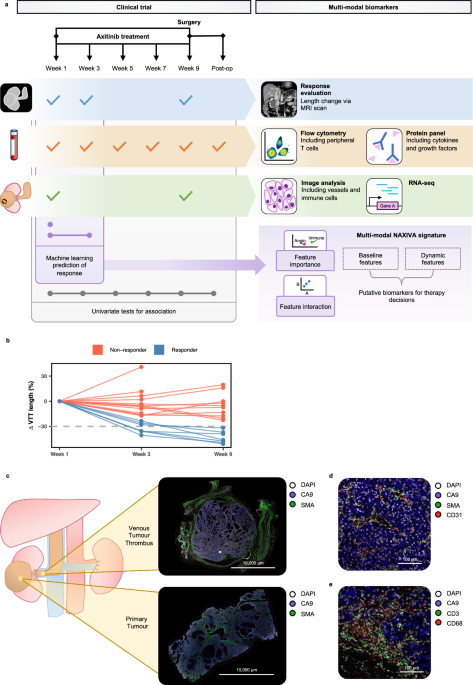
About 10-15% of cases of kidney cancer develop venous tumour thrombus (“VTT”) – an unusual phenomenon where the kidney tumour invades the major vessels of the abdomen, potentially threatening the liver and heart. These patients can be cured by extensive surgery; however, the operation carries significant risks. The NAXIVA trial (Phase II Neoadjuvant Study of Axitinib for Reducing Extent of Venous Tumour Thrombus in Clear Cell Renal Cell Cancer With Venous Invasion, NCT03494816) investigated the strategy of giving drug treatment to shrink the VTT before the operation. In NAXIVA, 20 patients were given up to 8 weeks of axitinib, a tablet treatment that disrupts signals the tumour needs to make new blood vessels. NAXIVA showed that there was a significant reduction in VTT in 35% of the patients that took part in the study, a positive result. The results of NAXIVA were previously reported in the British Journal of Cancer (Stewart et al, 2022). Both in this preoperative setting, and in the wider management of kidney cancer, a key question is why some patients respond to treatment and others do not. We aimed to explore this through the scientific analysis of samples collected during the NAXIVA trial, as recently reported in Nature Communications.
We found that patients who had a good response to treatment had a high density of blood vessels in the tumour at the start of the trial, and this had reduced significantly when the tumour was examined after surgery. We also found that patients who did not respond as well had higher levels of immune activation in the bloodstream. This fits with findings in advanced kidney cancer, where it has been observed that patients with blood vessel-rich tumours tend to respond well to these blood vessel directed therapies, whereas patients with immune activation tend to have more aggressive tumours and do better on immune-directed therapy.
We also identified that in patients who responded well, the levels of placental growth factor, a key signalling factor for blood vessel development, increased significantly in the blood stream early on treatment. This could be used as an early indicator that patients are responding well to treatment. A historic problem in developing blood-based predictors of response is the variation in individuals’ biology; by looking at these dynamic markers where we compare a reading taken early on treatment to one taken just before we may be able to account for natural variation and improve our predictions.
For each patient, we collected over 60 pieces of data before treatment, and many more during the treatment course. We used a machine learning approach to model the treatment response and identify biomarkers predictive of that response. Despite a relatively small number of patients, we were able to develop a successful model through “leave-one-out” approach, where the model is trained many times, each time with a different patient “left-out” to test it (i.e. to predict the “left-out” patient’s response), while the rest of the data is used to train the model (i.e. define its parameters). Our model first identifies the most important biological markers associated with treatment response among all markers recorded in the study and then predicts an individual patient's response based on those selected markers. The model repeatedly selected specific immune- and blood vessel-related markers and achieved good accuracy in predicting response. We extended the model to predict response using markers which were collected after treatment began, which improved the model accuracy. The machine learning approach is powerful as it allows us to take diverse, multi-dimensional information into account to predict response, and to understand not only which factors were involved, but also their relative importance.
There were several challenges in successfully completing the study. Clinical trials around the time of surgery are difficult to accomplish, as they require excellent coordination between specialists: surgeons, oncologists, radiologists, pathologists and the wider clinical research team. This team-based approach was also essential for the scientific analysis. We received excellent support from the core facilities on the Cambridge Biomedical Campus - for example the Core Biochemical Assay Laboratory, who were able to measure the level of cytokines in very small volumes of plasma, allowing us to gain maximum information from valuable blood samples. A particular challenge was getting good quality RNA data from paraffin preserved tumour samples, the Genomics Core Facility at the Cancer Research UK Cambridge Institute did excellent work in validating a new approach so that we could get RNA data from our samples, which added valuable insights to the study findings.
The involvement of Dr Mireia Crispin-Ortuzar’s computational oncology team at the Early Cancer Institute - including Rebecca Wray and Hania Paverd - was transformative in our understanding of the study data. Their insights from the transcriptomic and machine learning approaches allowed us to gain a detailed understanding of the features driving response in each patient. The use of machine learning to maximise the insights from clinical trial datasets will be central to the success of future studies.
The lessons from NAXIVA are being applied to ongoing pre-surgical clinical studies being run in Cambridge, including the WIRE study (WIndow of Opportunity Clinical Trials Platform for Evaluation of Novel Treatments Strategies in REnal Cell Cancer, NCT03741426). In WIRE, patients receive one cycle of treatment before planned nephrectomy. Response is tracked by dynamic contrast enhanced MRI scanning. Translational sampling is embedded in the design, so we can understand the mechanisms of response in each patient. Integrating different data streams using advanced computational approaches will be central to the analysis.
In the routine management of kidney cancer, the neoadjuvant treatment approach has not been widely adopted, largely because most evidence is based on Phase II studies such NAXIVA. With this in mind, the International Neoadjuvant Kidney Cancer Consortium (INKCC) has been set up with the aim of promoting well-designed prospective randomised studies that investigate the question of neoadjuvant treatment in kidney cancer.
The NAXIVA study not only sheds light on the biological factors influencing response to pre-operative axitinib but also underscores the critical role of advanced computational methods in analysing complex clinical trial data, informing the design of future studies. We are very grateful to the patients and families who took part in NAXIVA, and we hope that the results of this study will lead to improved outcomes for patients with kidney cancer in future.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in