Behind the Paper: Developing a Smarter CRISPR Tool with RNA-Sensing Capabilities
Published in Protocols & Methods, Cell & Molecular Biology, and Genetics & Genomics
The CRISPR Challenge: Precision and Control
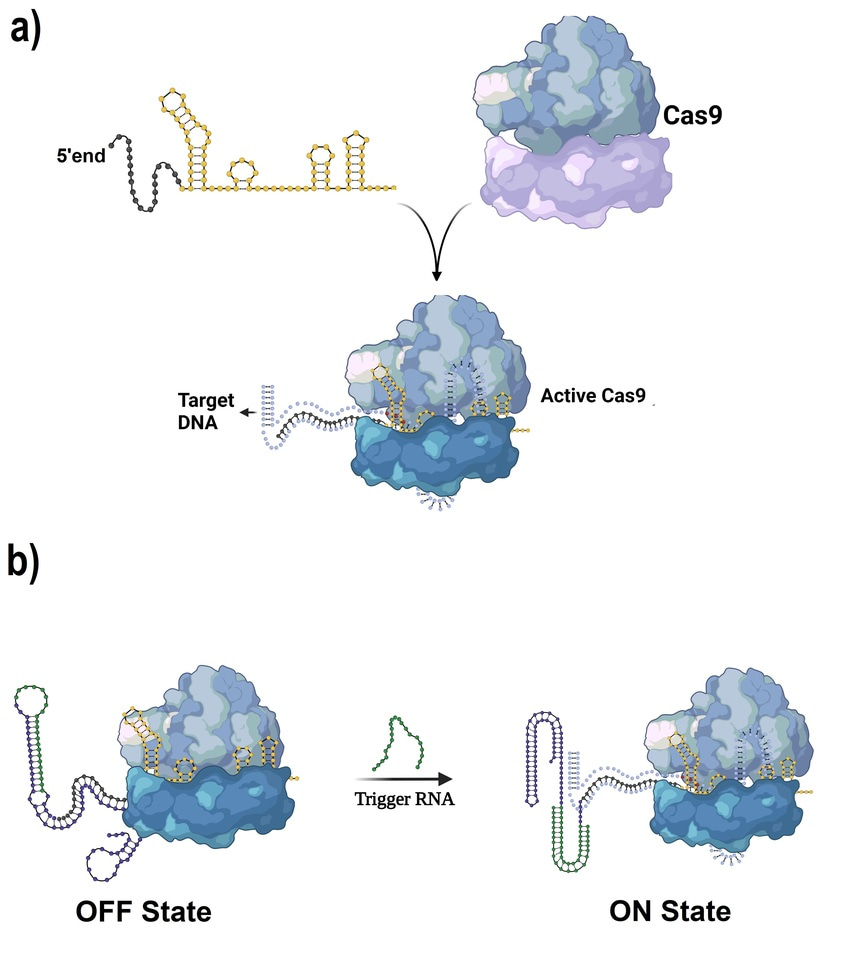
CRISPR/Cas9 systems have revolutionized the way we edit genes, offering unparalleled accuracy. However, a persistent issue with CRISPR is its continuous activity, which can sometimes lead to off-target effects—unintended edits that can cause cell damage or unexpected results. This is where our innovation comes in. Instead of allowing the CRISPR system to operate continuously, we designed it to remain in an "OFF" state until a specific RNA trigger is present. This creates a safer and more precise approach to gene editing.
IngRNA: A Smarter Gene Editor
At the core of our platform is a synthetic circuit that employs toehold switches—specially designed RNA sequences that respond to trigger RNA molecules. The system remains dormant until it detects a specific RNA, at which point it activates the CRISPR mechanism to edit the target gene. For this study, we focused on targeting the luciferase gene as a model, but the potential applications of this technology extend far beyond.
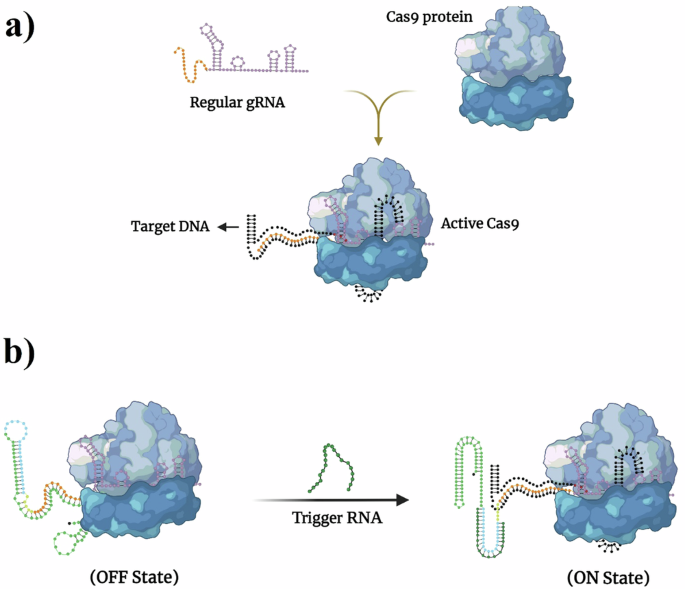
The IngRNA system incorporates two key toehold switches and an inhibited Crispr RNA (CrRNA) sequence. This design ensures that in the absence of the trigger RNA, the CrRNA remains inactive. Once the trigger RNA binds to the first toehold site, it initiates a cascade reaction that releases the CrRNA to perform its function—cutting the target gene with precision.
Why RNA Sensing?
In traditional CRISPR setups, the guide RNA is always active, which can lead to unwanted cuts in the genome. By incorporating RNA sensing into the system, we've given CRISPR a kind of "intelligence." The system only activates when it detects a specific RNA sequence, which acts as a signal that the target gene should be edited. This innovation not only enhances precision but also makes the system more responsive to the dynamic environment of a cell, allowing it to adjust its behavior based on real-time cellular conditions.
Results and Insights
Our experiments spanned several systems—from test tubes to bacterial cells to mammalian cells. In each case, we observed that the IngRNA platform successfully recognized trigger RNA and modulated gene activity. In bacterial cells, for example, we showed that the presence of a trigger RNA could activate the system to cleave the luciferase gene, effectively "switching off" the reporter gene's signal. When the trigger RNA was absent, the system remained in its off state, preventing any unintended gene editing.
Moreover, by enhancing the system to IngRNA2, we improved the platform’s stability, ensuring that it remains in the off state until the trigger RNA is present. This advancement reduces the risk of spontaneous activation, a common challenge in earlier gene-editing platforms.
Beyond Bacteria: Application in Mammalian Cells
We didn't stop at bacterial systems. One of the most exciting aspects of our research was testing the IngRNA platform in mammalian cells. We used human HEK293 cells and tested the platform with miR-16, an RNA molecule associated with apoptosis (cell death). Our results were promising—when the system detected miR-16, it activated the CRISPR function, targeting the luciferase gene and suppressing its activity. This ability to sense endogenous RNA molecules and adjust gene activity accordingly opens up exciting possibilities for therapeutic applications.
What’s Next?
The potential applications of this platform are vast. In cancer research, for example, IngRNA could be used to selectively target cancer cells by detecting RNA markers specific to tumors. In gene therapy, it could offer a safer way to edit genes by ensuring that CRISPR only activates in the presence of a disease marker. The ability to integrate RNA sensing with gene editing provides a powerful new tool for scientists working in synthetic biology, diagnostics, and therapeutics.
Reflecting on the Journey
Developing the IngRNA platform was both challenging and rewarding. From optimizing the toehold switches to ensuring stability in different cellular environments, the process required a deep understanding of both RNA nanotechnology and CRISPR biology. Our team's collaborative spirit and determination to solve real-world problems in gene editing kept us motivated throughout.
We hope that this research not only advances the field of gene editing but also inspires other scientists to explore new ways to make CRISPR even smarter and more precise.
For more details, you can find our full research article here [article link].
Follow the Topic
-
Communications Biology

An open access journal from Nature Portfolio publishing high-quality research, reviews and commentary in all areas of the biological sciences, representing significant advances and bringing new biological insight to a specialized area of research.
Related Collections
With Collections, you can get published faster and increase your visibility.
Signalling Pathways of Innate Immunity
Publishing Model: Hybrid
Deadline: Feb 28, 2026
Forces in Cell Biology
Publishing Model: Open Access
Deadline: Apr 30, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in