Cancer-Alterome, how literature resources contribute to the refined interpretation of cancer pathology ?
Published in Cancer and Computational Sciences
Cancer has long been a significant global health concern, posing a serious threat to human health and life. The occurrence and progression of cancer are often associated with complex regulatory events brought about by genetic alterations. Meanwhile, gaining a deeper understanding of the molecular mechanisms behind these regulatory events holds the promise of treating and overcoming cancer.
Our team has been dedicated to developing text-mining methods to capture regulatory events brought about by genetic alterations from literature, thus aiding in the elucidation of fine-grained disease mechanisms. The Cancer-Alterome[2] represents a milestone in this series of studies. This work relies on our previously developed AGAC[1] (Annotation of Genes with Alteration-Centric function changes) corpus. It defines eight categories of biological concepts spanning from molecular to cellular levels, including genes, mutations, molecular process activities, cellular process activities, diseases, and more. Additionally, it defines two types of relationships to compose alteration-centric biological regulatory events, e.g. “ThemeOf” relationships between genes and alteration, and “CauseOf” relationships between alteration and downstream events. The Alteration-caused Regulatory Events (GARE) in Cancer-Alterome are a continuation of this definition.
We are proud to introduce Cancer-Alterome (http://lit-evi.hzau.edu.cn/PanCancer), a finely curated resource of cancer pathology descriptions constructed from the scientific literature. In the creation of this resource, we utilized a series of mature text-mining methods to gather and process cancer-related scientific literature. Leveraging the AGAC corpus, we completed the definition and precise capture of GARE. Ultimately, data repositories and web services are provided to facilitate domain experts' utilization and further development of this resource (Fig. 1).
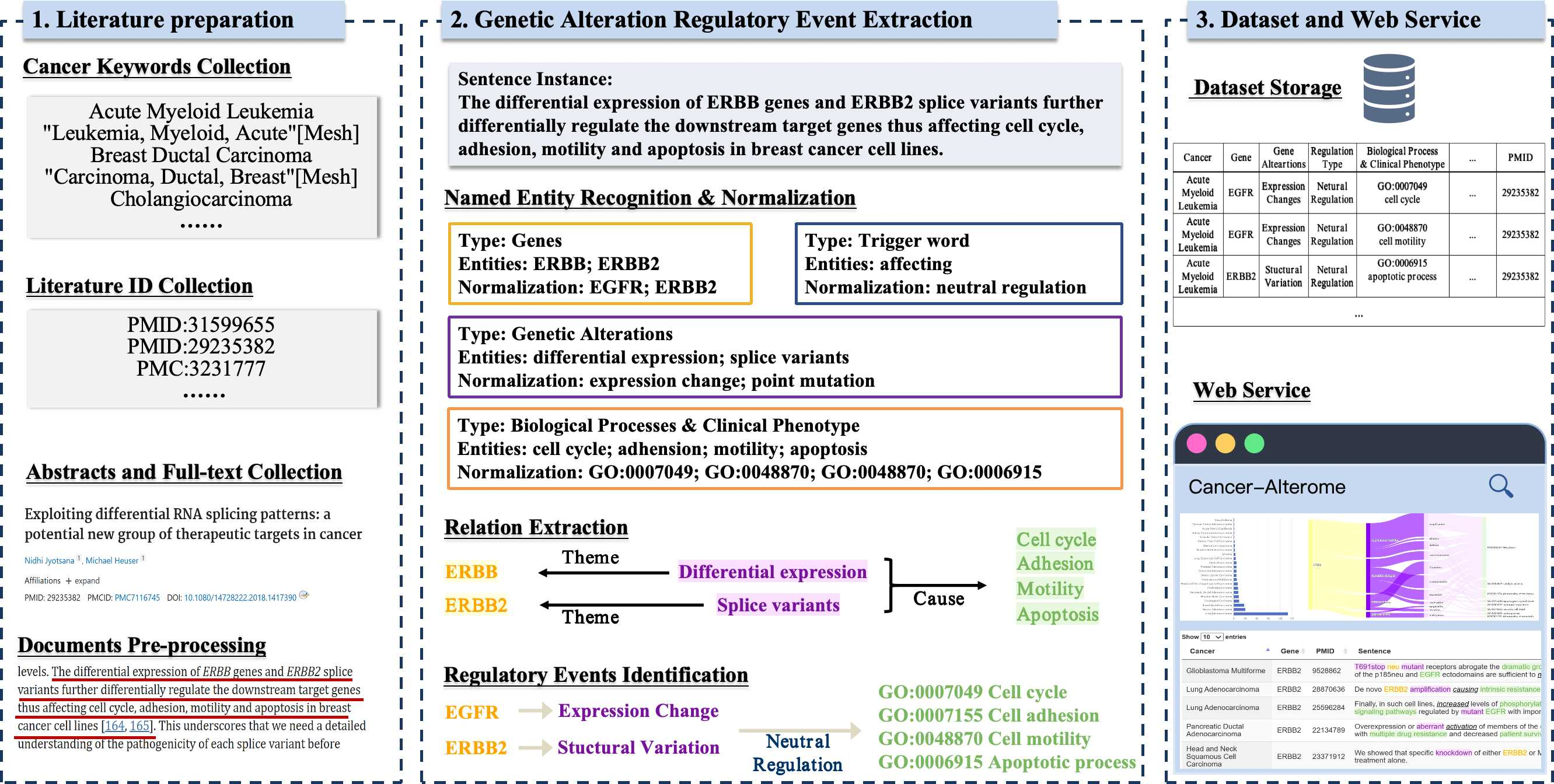
Figure 1. Data processing details of the pipeline with an example.
The Cancer-Alterome represents a pioneering effort in achieving such comprehensive and granular coverage of cancer literature to date. Specifically, Cancer-Alterome focuses on 32 pan-cancer types defined in TCGA, gathering a staggering 4,354K articles and ultimately extracting 16,581k records of GARE. On average, each cancer entails 521K regulatory events. These records encompass 21K human genes, 136K dbSNP-standardized genetic mutations, and descriptions of 20K genetic alterations. Moreover, all downstream events have been standardized to over 4K GO terms, 2K HPO terms, and 146K MeSH terms. Finally, Cancer-Alterome's web services (http://lit-evi.hzau.edu.cn/PanCancer) and data repository (https://github.com/YaoXinZhi/Cancer-Alterome) have been made available to the scientific community.
The downstream applications of Cancer-Alterome hold promising prospects, and our team has made meaningful attempts in this regard. On one hand, this resource can provide literature and statistical support for the mechanistic roles and biological correlations of key biomarkers in diseases, much like what Cancer-Alterome has achieved. On the other hand, the integration of mutation regulatory events described in the literature with multi-omics data holds the potential to further pinpoint crucial disease biomarkers, thereby facilitating knowledge discovery. In the work of 2021[3], in order to integrate heterogeneous mutation data, we proposed the "Gene-Disease Association Prediction through Mutation Data Bridging (GDAMDB)" pipeline and established a statistical generative model. This model can learn the distribution parameters of mutation associations and mutation types, and identify false-negative GWAS mutations that are supported by evidence representing functional biological processes in the literature but were not significant in conventional tests. Recently, our work[4] has further combined GARE knowledge with sequence analysis data, proposing a Bayesian deep learning model called PheSeq, which enhances and interprets association studies by integrating and perceiving phenotypic descriptions. This model also generates a vast dataset of association evidence, opening new possibilities for interpreting and exploring gene-disease associations.
Finally, we are delighted to share our work with the scientific community and domain experts in the prestigious journal, Scientific Data. We sincerely hope that this resource can provide valuable research groundwork and further insights for the community.
Reference
- Yuxing Wang, Kaiyin Zhou, Jin-Dong Kim, Kevin Cohen, Mina Gachloo, Yuxin Ren, Shanghui Nie, Xuan Qin, Panzhong Lu, Jingbo Xia*. An Active Gene Annotation Corpus and Its Application on Anti-epilepsy Drug Discovery. BIBM 2019: International Conference on Bioinformatics & Biomedicine. Page: 512-519, San Diego, U.S, Nov, 2019.
- Xinzhi Yao, Zhihan He, Yawen Liu, Yuxing Wang, Sizhuo Ouyang, and Jingbo Xia*. Cancer-Alterome: a literature-mined resource for regulatory events caused by genetic alterations in cancer. Scientific Data. 2024, 11:265. DOI: 1038/s41597-024-03083-9. (https://www.nature.com/articles/s41597-024-03083-9)
- Kaiyin Zhou#, Yuxing Wang#, Kevin Bretonnel Cohen, Jin-Dong Kim, Xiaohang Ma, Zhixue Shen, Xiangyu Meng, Jingbo Xia*. Bridging Heterogeneous Mutation Data to Enhance Disease-Gene Discovery. Briefing in Bioinformatics, 2021, bbab079.
- Xinzhi Yao, Sizhuo Ouyang, Yulong Lian, Qianqian Peng, Xionghui Zhou, Feier Huang, Xuehai Hu, Feng Shi, Jingbo Xia*. PheSeq, A Bayesian Deep Learning Model to Enhance and Interpret the Gene Disease Association Studies. Genome Medicine, 2024, 16:56. DOI: 10.1186/s13073-024-01330-7.
Written by Zhihan He, Xinzhi Yao, and Jingbo Xia.
Follow the Topic
-
Scientific Data

A peer-reviewed, open-access journal for descriptions of datasets, and research that advances the sharing and reuse of scientific data.
Related Collections
With Collections, you can get published faster and increase your visibility.
Data for crop management
Publishing Model: Open Access
Deadline: Apr 17, 2026
Data to support drug discovery
Publishing Model: Open Access
Deadline: Apr 22, 2026





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in