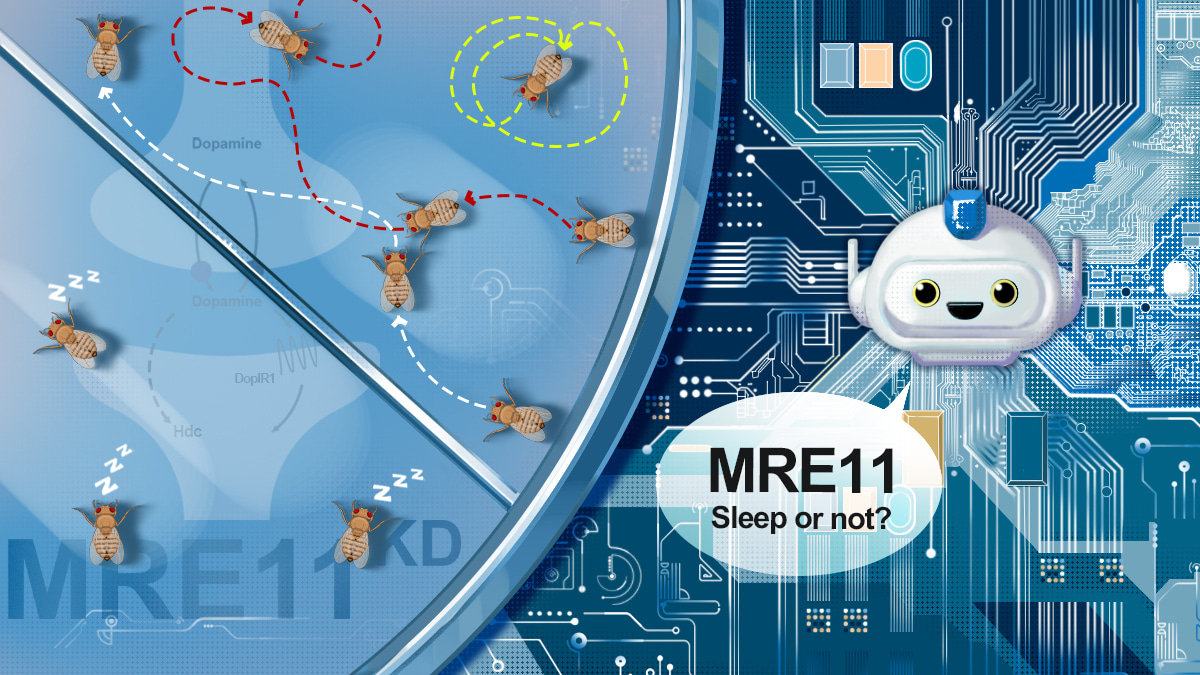
Sleep is believed to be quite adaptive and prone to be influenced by environmental factors (1). Since animals rarely act in isolation, a key environmental factor is social signal. However, due to technological limitations, the great majority of previous studies measured the sleep of an animal in an isolated condition, which deviates from the natural habitat of these animals. This lead us to suspect that perhaps if we could assess the sleep of each animal living in a group, the quality and quantity of sleep would be different from that of isolated individuals. We contemplated this idea in 2019 and put it into practice shortly after. Luoying’s laboratory has been studying sleep and circadian rhythm in fruit flies, and thus we developed a multi-object video-tracking system for monitoring fly locomotor activity. Using this system, we can assay sleep, locomotion and social activity of each fly with high accuracy (greater than 98%) in a group consisting of 5 or fewer individuals. We found that sleep duration is shortened by approximately 200 minutes when there are 3 or more flies in a group, which is caused by inhibition of both sleep initiation and maintenance. These changes are substantial indeed, but not very surprising. It is expectable that when there are other animals present, sleep would more likely to be disturbed by others.
We next conducted a genome-wide screen in our video-tracking system. We expressed 6,885 RNA interference (RNAi) constructs pan-neuronally that target 5,588 genes, and identified 285 genes that function to promote sleep. What surprised us was that only 20 of the known sleep-regulating genes were identified by our screen. This could certainly be due to insufficient silencing of the target genes, while a more fascinating possibility is that the molecular mechanism regulating sleep under group condition is rather different from that of isolated condition. The latter notion is supported by genes such as mre11 (a gene involved in DNA damage repair and telomere protection), which exhibit strong sleep-promoting effects only when there are other flies present. Characterizing the mechanism by which mre11 promotes sleep, however, was quite a challenging task. We knocked down mre11 in various brain structures and cell types that are known to be involved in sleep regulation, but failed to observe the short-sleep phenotype. This implies that the neural circuitry that controls sleep under group condition may be distinct from that of isolated condition. Pharmacological experiments provided a clue that mre11 may promote sleep via dopaminergic signaling. At this time, the project came to a bottleneck.
On the night of January 31, 2023, Yu read in the news regarding the release of a new update of Generative Pre-trained Transformer (GPT), ChatGPT or GPT 3.5. When GPT 3 was released in 2020 (2), Yu, who is a bioinformatician, and his group devoted much effort into employing GPT 3 to resolve biological questions but failed. Later in 2021, one of Yu’s friends and a leading bioinformatician, Dr. Han Liang, published a paper reporting a computational platform, DrBioRight, which implemented a human-AI dialogue interface to enable computers to perform various data analysis tasks (3). DrBioRight displayed the ability of machine understanding of human languages, and Dr. Liang suggested that NLP might be useful in performing bioinformatic tasks. Yu decided to test whether ChatGPT can be useful in resolving biological questions, and perhaps help advance our sleep project.
After testing for 4 days, we found that the knowledge in ChatGPT can be efficiently elicited by a cutting-edge technology, prompt engineering. Initially, we observed that if the prompt was casually written, the response of ChatGPT was also casual, and AI hallucination frequently occurred to output incorrect answers. Thus, we realized that the prompt needs to be carefully designed, and for querying a detailed problem, some necessary background knowledge should be included in the prompt to elicit the correct knowledge. Using the standard prompting strategy, we first performed a genome-wide interpretation in Drosophila melanogaster by obtaining 128,382 prompt-generation pairs, and obtained 12.5%, 13.8%, and 10.2% of the fly protein isoforms interpreted to be involved in sleep, locomotor and social activity regulation, respectively, with a low false positive rate of ~7%. This demonstrates the superiority of ChatGPT in gathering and comprehending scientific knowledge.
More importantly, we observed an intriguing feature of ChatGPT during the genome-wide interpretation. We found that the chain of thought (CoT) could be elicited in a step by step manner, and the interpretation can be reasoned in a human-like logic. This means based on the existing knowledge, ChatGPT can make predictions regarding the functional associations of genes. For example, if gene A is known to regulate gene B in one context, while gene B is known to regulate gene C in another context, then ChatGPT predicts that gene A may regulate gene C. To make use of this feature, we compiled 86 top candidate genes from our genetic screen, including 19 genes known to be involved in regulating sleep, locomotion and/or social activity. The CoT prompting strategy was used to reason potential regulations or associations of 86 * (86-1)/2 = 3,655 gene pairs. From the results, 139 pairs of potential regulations or associations were acquired via machine reasoning, and 103 (74.1%) of these answers were supported by the literature. Based on machine reasoning, mre11 potentially regulates two genes encoding dopamine receptors (Dop1R1 and DopEcR) and histidine decarboxylase (Hdc) to promote sleep. We first assessed whether the expression of these three genes are altered in mre11 RNAi flies and found that Hdc mRNA level is reduced. Our screen results showed that silencing Hdc leads to decreased sleep, and thus Hdc may indeed mediate the sleep promoting function of mre11. Moreover, we found that treating flies with Dop1R antagonist can partially reverse the sleep phenotypes (more precisely the sleep initiation impairments) caused by mre11 deficiency. This suggests that mre11 may promote sleep at least in part via Dop1R1. We shall further validate these predicted targets and investigate the underlying mechanism in our future research. All in all, this reveals the unprecedented ability of ChatGPT to help bridge the gap in human knowledge and facilitate scientific discoveries.
Notably, this may be the first time that ChatGPT is used for resolving a complex biological problem, and such a human-AI interactive paradigm may be extended to other fields beyond biology.
This work lead to the awarding of a grant from Special Key Projects of National Natural Science Foundation of China ( “AI analysis of the logic and structure of molecular languages in life processes”), with grant numbers of 32341020 and 32341021. Yu and Luoying are joint principle investigators on this grant entitled “AI analysis and application of molecular languages in physiological and pathological processes”.
Related references:
- Allada, R. and Siegel, J.M. (2008) Unearthing the phylogenetic roots of sleep. Curr Biol, 18, R670-R679.
- Brown, T.B., Mann, B., Ryder, N., Subbiah, M., Kaplan, J., Dhariwal, P., Neelakantan, A., Shyam, P., Sastry, G., Askell, A. et al. (2020) Language Models are Few-Shot Learners. 10.48550/arXiv.2005.14165.
- Li, J., Chen, H., Wang, Y., Chen, M.M. and Liang, H. (2021) Next-Generation Analytics for Omics Data. Cancer cell, 39, 3-6.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in
Very instructive!llm for efficient imaging-based cellular analysis is on my to-do list!