Chlamydia in human organoids and the origin of fallopian tube disease
Published in Microbiology

As infections frequently remain asymptomatic, providing no opportunity for timely medial intervention, they can become chronic. Clinical practice and epidemiological data strongly suggest that such protracted chronic infections and recurrent episodes can cause scarring of the fallopian tubes, which may not merely prevent conception but also increases the risk of ectopic pregnancy. In addition, mounting epidemiological evidence suggests that C. trachomatis infection increases the risk of developing ovarian cancer, which in most cases originates from the distal end of the fallopian tube.
Despite its widespread impact on women’s health, however, there has been very limited insight into the mechanisms through which C. trachomatis interacts with the fallopian tube epithelium, mostly due to the absence of adequate experimental models. As the primary focus of interest in our lab at the Max Planck Institute for Infection Biology in Berlin has always been the influence of pathogens on the host cells and tissue, we recognized quite early on that one of the major challenges in understanding its role in tubal pathology was the need for a suitable in vitro model system. Our initial exploratory study with ex vivo infection of native fallopian tube tissue pieces indicated that paracrine signaling pathways played an important role in the epithelial response to invading C. trachomatis, highlighting the need to study pathogen-host interaction in a system with intact epithelial polarity and cell-cell communication (Kessler et al 2012).
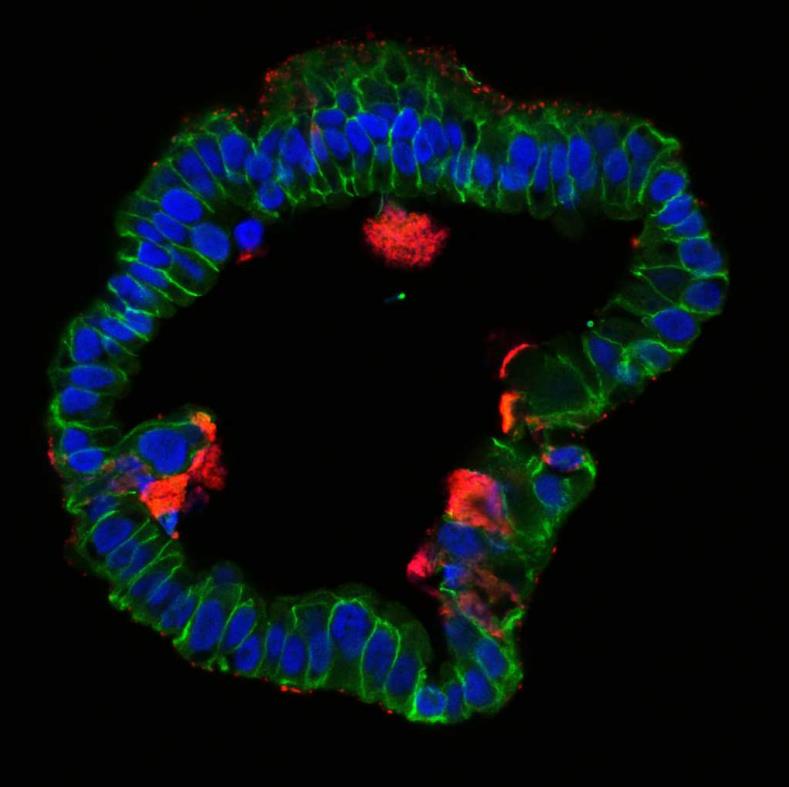
With all these factors in mind, establishment of long-term stable organoid cultures in our lab (Kessler et al 2015) using clinical samples of human fallopian tube proved to be a “game changer”, and provided a unique opportunity to study numerous open question about the impact of Chlamydia on this epithelium. To our great surprise, infection of organoids revealed exciting novel aspects of the host defense against the pathogen and differed fundamentally from the course of infection seen previously in immortalized cell lines. We discovered that the organoid epithelium, while readily susceptible to invading Chlamydia, has potent mechanisms to contain the infection by efficiently shedding inclusions and infected cells.
Importantly, the organoids continued to grow, even as the ongoing reinfection cycles within the system recreated a chronic infection that could be analyzed under controlled in vitro conditions. Our comprehensive analysis of phenotypic and global gene expression changes over the course of 9 months revealed that C. trachomatis infection affects differentiation of the epithelial cells and increases organoid forming efficiency. Strikingly, infected organoids showed an increase in CpG hypermethylation in a pattern that is known to be associated with cellular ageing. This exciting finding is the first direct evidence that this pathogen has the capacity to affect the epigenome and thereby influence epithelial fate beyond the resolution of infection.
Numerous open questions remain to obtain a complete understanding of the complexity of Chlamydia infections, such as the mechanisms underlying bacterial clearance and reinfection, the crosstalk between inflammatory response and regulation of stemness and the potential impact of C. trachomatis on cellular transformation in ovarian cancer. We are confident that this novel infection model provides a great basis for fundamentally improving our knowledge about Chlamydia-driven tubal pathology in the near future.
References
Kessler M, Hoffmann K, Fritsche K, Brinkmann V, Mollenkopf HJ, Thieck O,Teixeira da Costa AR, Braicu EI, Sehouli J, Mangler M, Berger H, Meyer TF (2019) Chronic Chlamydia infection in human organoids increases stemness and promotes age-dependent CpG methylation. Nat Commun. 2019 Mar 18;10(1):1194
Kessler M, Hoffmann K, Brinkmann V, Thieck O, Jackisch S, Toelle B, Berger H, Mollenkopf HJ, Mangler M, Sehouli J, Fotopoulou C, Meyer TF (2015) The Notch and Wnt pathways regulate stemness and differentiation in human fallopian tube organoids. Nat Commun 6: 8989
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in