CLOOME: contrastive learning unlocks bioimaging databases for queries with chemical structures
Published in Protocols & Methods, Cell & Molecular Biology, and Computational Sciences
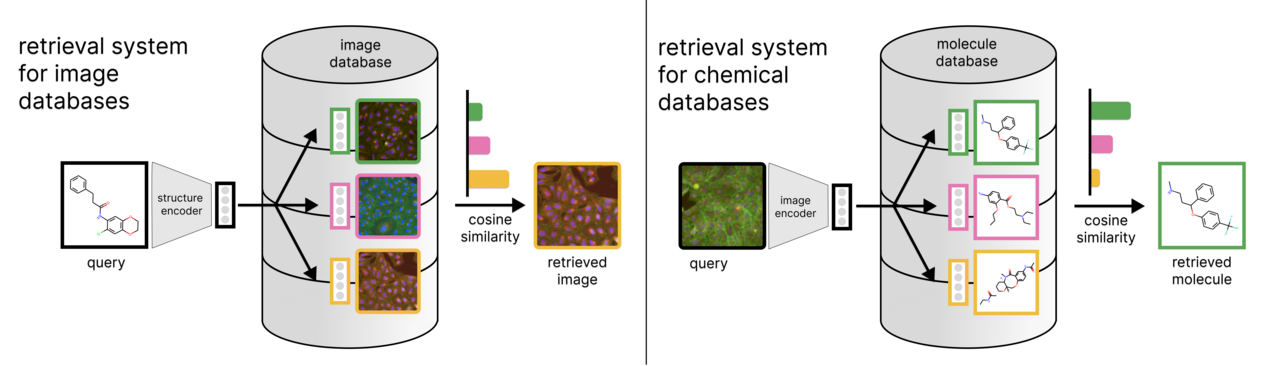
Biological and chemical databases enable much of the research that is produced these days. Therefore, finding ways to extract specific, relevant information from them can greatly optimize how scientists have access to this information and, ultimately, accelerate the pace at which research findings are made. So, over the years, different querying mechanisms have been curated for different data types in the life sciences. Both in sequence and structure databases, such as RefSeq or PDB, users are given the possibility to find similar entries to their queries using methods particularly designed for it, such as BLAST or structure similarity search via shape descriptors.
However, bioimaging databases, at the moment, can only be queried by text annotations (e. g. a molecule identifier, such as an InChIKey). This query would yield the exact image of cells that were treated with the query molecule, but would not give us a ranking of other similar samples. To overcome this limitation, we developed CLOOME, a method that allows querying bioimaging databases with molecular structures to obtain a ranking of images in the database that potentially show the same phenotypic effect as that of the query molecule. Conversely, CLOOME also allows querying a chemical database with an image of a certain phenotype.
The methodology behind CLOOME’s ability to map bioimages and chemical structures is contrastive learning. Specifically, CLOOME is based on the contrastive learning methods CLIP and CLOOB, which we adapted from natural images and text pairs to microscopy images and molecule pairs. This method works in such a way that both the images and the molecules are converted into vectors that are embedded into the same space, and then the image and molecule vector pairs that belong together are pulled closer together, while the non-matching vector pairs are pushed far apart. Therefore, molecular structures can query a bioimaging database by selecting images that are nearby in the embedding space, and vice versa.
In our paper, we found the following:
1. CLOOME performs far better than the baselines for chemical and bioimaging database retrieval.
We tested the ability of CLOOME to correctly retrieve the matching sample from a chemical or bioimaging database. For the chemical database retrieval, given a microscopy image of cells treated with a certain compound, the task is to select the correct chemical structure from a set of thousands of candidate structures. Since cells often do not show any or only subtle phenotypic changes, this is a very challenging task for human experts. For the bioimaging database retrieval, given a molecule, the task is to find the matching microscopy image, which is similarly difficult.
This image-based retrieval task can also be understood as a bioisosteric replacement task: bioisosteres are molecules for which an atom or a set of atoms are different, but present similar biological properties. With this experiment, we evaluate the ability of CLOOME to correctly rank the matched molecular structure given the corresponding image. Other high-ranked structures could be potential bioisosteres, which makes this experiment a proxy for the bioisosteric replacement problems.
2. CLOOME can be used to predict compound activities in bioassays
One asset of contrastive learning methods is that, as they are trained in a self-supervised way, they produce a robust representation of the data, so that this representation can be transferred to different tasks. In this case, we tested whether the representations learned by CLOOME are transferable to predicting assay bioactivity. We used the representations of the pre-trained encoders and fitted a logistic regression model to predict the given labels.
We compared our method to other fully supervised methods like convolutional neural networks (CNNs) trained in a fully supervised way, as well as to logistic regression fitted to CellProfiler features, which could also be considered as a self-supervised method. CLOOME embeddings outperformed CellProfiler features and were comparable to fully supervised methods.
3. CLOOME also has the potential for zero-shot classification of molecules and mechanism of action (MoA).
Finally, we also assessed whether the CLOOME image embeddings could be used to correctly predict the mechanism of action (MoA) or the molecule of a given image. This could be considered a zero-shot classification task, as the model was not trained to predict MoAs. To perform this experiment, one image for each of the MoAs in a hold-out test set was randomly selected, which means that each unseen MoA class is represented by a single image. We chose this setting to simulate a situation where we have a set of molecules for which we know its mechanism of action and for which we have available images, and we want to classify another set of images.
Then, samples from this set as well as samples corresponding to both the same molecule and plate were removed from the full test set, to ensure that the classification was not due to plate effects.
Results show that CLOOME presents better performance than GapNet and CellProfiler features in molecule and mechanism of action prediction
In summary, we presented CLOOME as a powerful cross-modal retrieval system between chemical structures and bioimages. This, for the first time, allows querying bioimaging databases by chemical structures or ranking them by similarity to a given bioimage or chemical structure.
We also showed that the CLOOME embeddings are transferable representations. This opens the possibility to re-use the learned representations for activity or mechanism of action prediction tasks.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in