Collaborative science bridges tiny streams and massive rivers, by connecting molecular processes to macroecological patterns
Published in Earth & Environment, Ecology & Evolution, and Microbiology
Explore the Research
A functional microbiome catalogue crowdsourced from North American rivers - Nature
GROWdb defines US river microbiomes at the genome level.
Microbes are everywhere and impact nearly every natural process we care about, from human health to the movement of carbon throughout the planet. The composition and function of microbial communities have been studied in many environmental systems, such as soils and the oceans, but microbes in streams and rivers had yet to be systematically explored.
The microbes in streams and rivers perform many functions; some are beneficial (e.g., keeping the water clean and cycling nutrients) and some are less so (e.g., generating methane, a potent greenhouse gas). This is the first study with sufficient data quantity and data type diversity and standardized collection, to be able to explore general patterns of microbial function across streams to large rivers. Being able to generalize patterns is critical to anticipate how changing conditions may impact these systems, and inform decisions on how to adapt or alter the outcome. But learning the rules that govern microbes is challenging, especially across a wide range of environmental gradients, from mountain headwaters that start as small creeks to the raging Mississippi River.
Our journey started with a more technical question: How do we connect detailed molecular processes across large scales, while still generating knowledge that’s relevant to many different scientific questions? The answer - for US river microbiomes, at least - lies in 7 years of effort, and input from more than 100 collaborative groups.
In 2017, it was clear we needed a new way of doing science. Heavily instrumented field sites were excellent for conducting deep mechanistic studies of hydrology and biogeochemistry, but those results may or may not be transferable to other locations/watersheds with varying environmental factors. We formed the Worldwide Hydrobiogeochemistry Observation Network for Dynamic River Systems (WHONDRS, pronounced as ‘wonders’; https://www.pnnl.gov/projects/WHONDRS) as a consortium based on participatory science. Co-led by Stegen and Amy Goldman (Pacific Northwest National Laboratory), 100s of individuals across 100s of organizations provided guidance on how to collect data and what data are most useful, and generated samples and data that have led to numerous collaborative publications. However, while WHONDRS researchers collect physical, chemical, and biological samples in and around streams and rivers, current analyses focus on organic matter chemistry. Publication co-corresponding author Kelly Wrighton saw an opportunity to add a microbiome component, with the goal of creating a Genome Resolved Open Watersheds database (GROWdb).
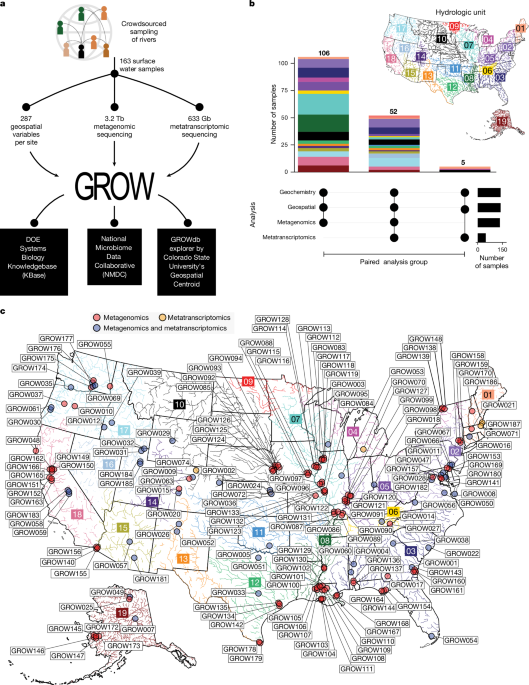
In 2020, Wrighton and lead author Mikayla Borton went beyond the WHONDRS consortium, targeting a broader set of collaborators interested in understanding the role of microbes in rivers. This consortium was awarded a Community Science Program (CSP) project by the Joint Genome Institute (JGI), a U.S. Department of Energy user facility, one of the first of JGI’s extra-large sequencing allocations, with >1000 total samples for DNA and RNA sequencing. The resulting GROWdb contains a reference database of more than 3,000 microbial genomes from about 100 rivers across North America. Across watersheds, the microbial gene expression data, which signifies how microbes are responding to their respective environments, indicates that our impact on rivers can be detected in microbial behavior. Perhaps a generalizable signal for determining contaminants and overall river health, and for detecting and monitoring changes over time!
Finally, in our commitment to the WHONDRS model of open science, all sample information and data generated as part of the GROWdb have been made FAIR (findable, accessible, interoperable, and reusable) at a variety of locations. The National Microbiome Data Collaborative (NMDC, https://microbiomedata.org) data portal provides access to the GROW project as part of a larger microbiome data integration hub. This includes the JGI analyses as linked metagenomic and metatranscriptomic data. The published river microbiome datasets are also available through the DOE Systems Biology Knowledgebase (KBase, www.kbase.us). KBase enables immediate reuse of GROWdb data, as users can quickly access and add GROW samples/data to their own analyses for comparison via the KBase platform. With no need to download and format data or load analysis packages and resources, saving researchers a lot of time, and free computing power. Finally, WHONDRS worldwide river sampling data are available at the Dept of Energy’s environmental systems science data repository, ESS-DIVE (https://data.ess-dive.lbl.gov/portals/WHONDRS/Data).
In the long run, we hope this enables non-microbiome scientists to start using these data. River microbiomes are important processes in our landscapes, and better representation will improve our capacity to anticipate and adapt to future environmental change.
As Borton says, “It takes a village to study across landscapes. Rivers are a system that can help us start to do that.”
The most exciting part? Borton et al. were recently awarded an even larger JGI sequencing grant focused on wetland soils around the globe. And we’ll keep making all the data openly available!
Follow the Topic
-
Nature

A weekly international journal publishing the finest peer-reviewed research in all fields of science and technology on the basis of its originality, importance, interdisciplinary interest, timeliness, accessibility, elegance and surprising conclusions.


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in