Combining metagenomics and network analyses identifies microbial interactions and their mechanism
Published in Microbiology
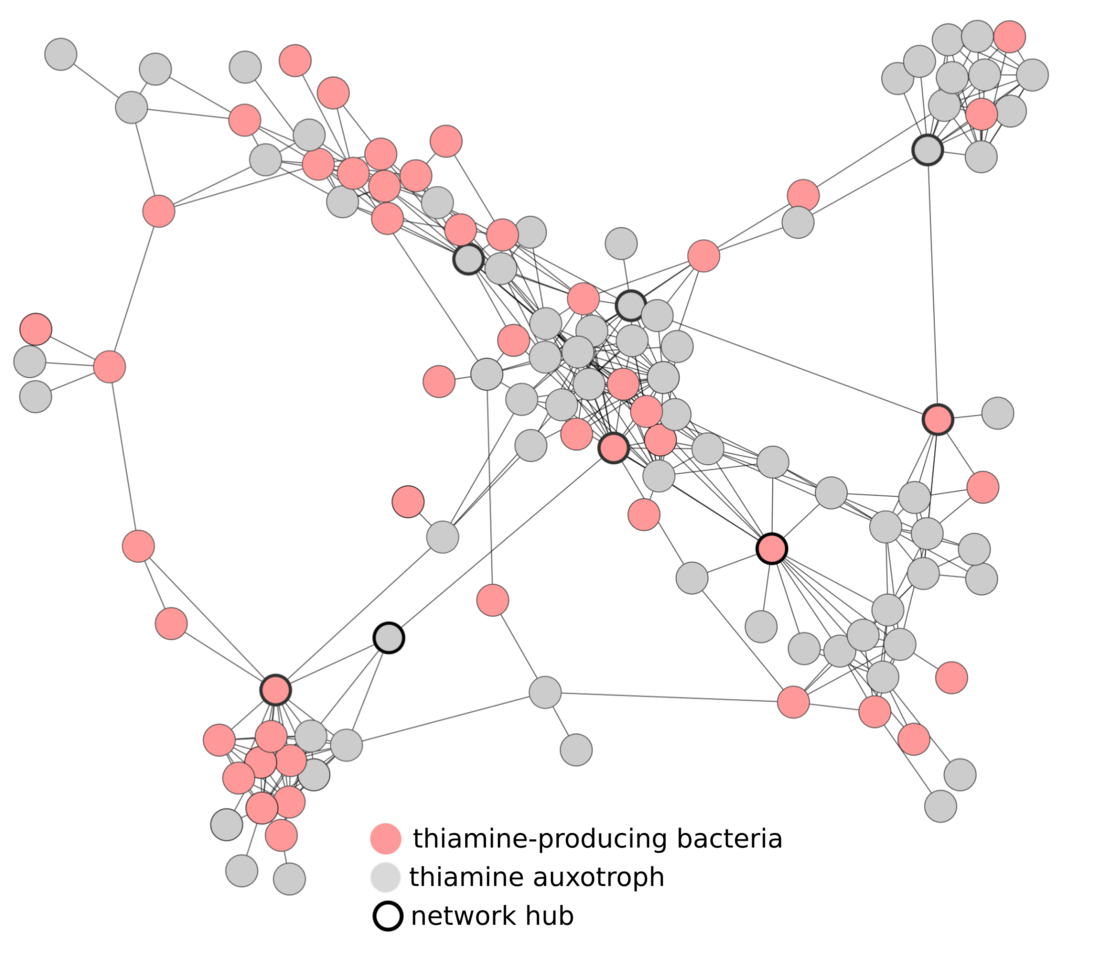
Bacteria seldom live alone, instead, they live within microbial communities where they exchange goods such as amino acids, nucleotides and vitamins. Exchanging these compounds reduces the number of compounds an organism needs to synthesize, and frees up time and energy for other important processes. Exchanging these metabolites can therefore be highly advantageous and is common across microbial communities. We can be sure that vitamins are commonly exchanged among bacteria because many vitamins are essential for microbial growth yet a large proportion of bacteria don’t encode the genes necessary for their production (Figure 1).

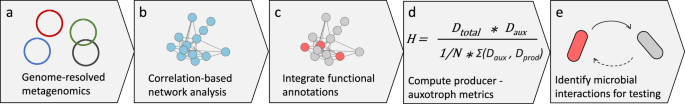
We supposed that organisms unable to produce particular vitamins may rely on specific vitamin-producing bacteria and we would, therefore, be able to observe this dependence across many samples by constructing correlation-based networks. To do this, we used metagenomic data associated with a range of bioreactors operated under 92 similar but varied conditions. We used metagenomic data to assess each microorganism’s genetic potential and determined whether each organism was able or unable to produce particular vitamins. We then overlaid this information onto the correlation network (Figure 2).

Considering the number of connections between vitamin-producing and auxotrophic bacteria (unable to produce a vitamin), we could estimate how important each organism within the network may be for the production of a given vitamin (Figure 3). Using this approach we identified a number of organisms that we predict to be important for supplying several B vitamins including thiamine, riboflavin, pantothenate and biotin to dependent organisms in their communities.
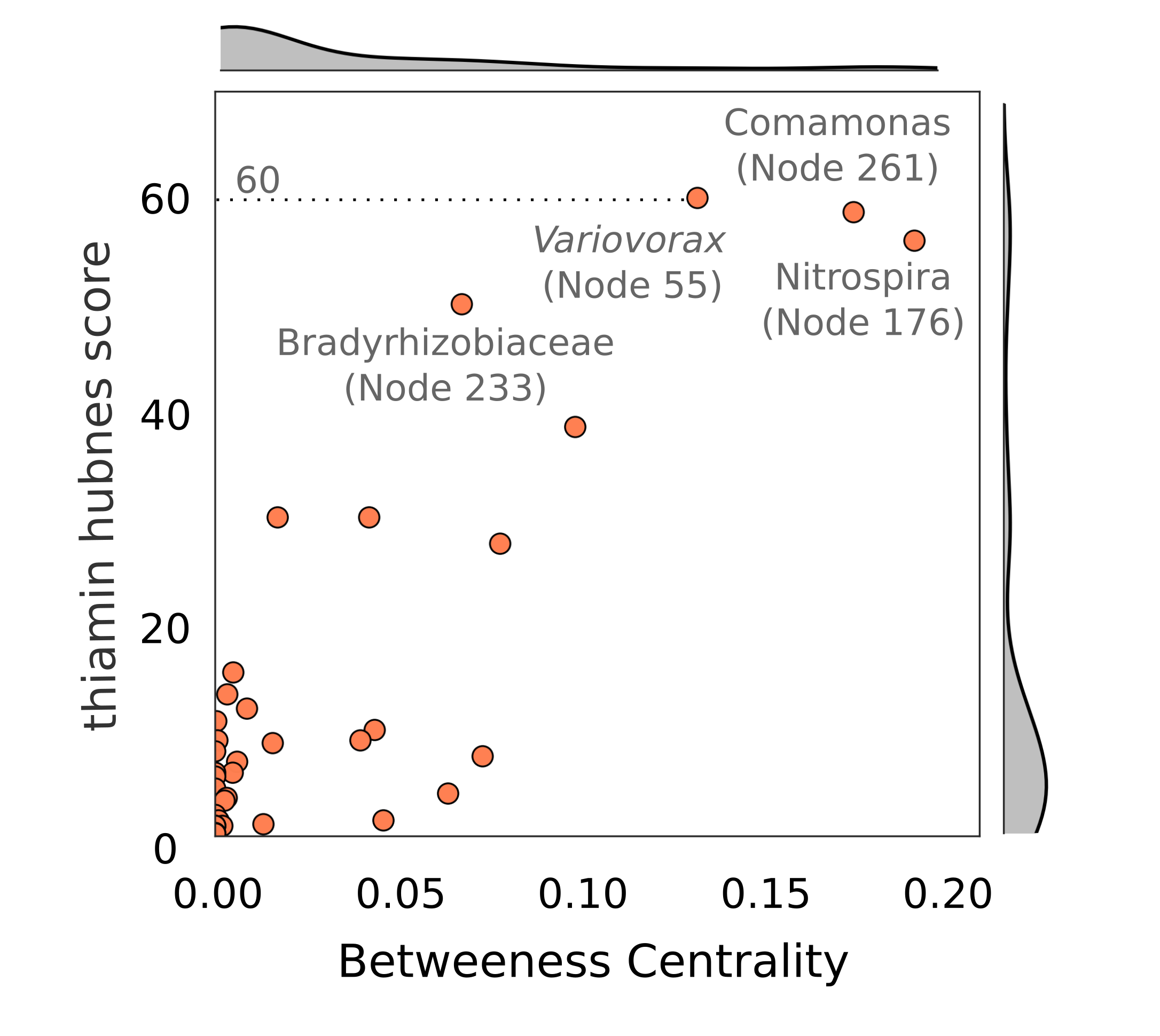
We then isolated many organisms from bioreactors performing thiocyanate degradation, including the predicted thiamine-producer, Variovorax. We first wanted to confirm that co-culturing Variovotrax with a thiamine auxotroph would boost the growth of the auxotroph. During our isolation campaign, we isolated a yeast, Cryptococcus, which was only able to grow in minimal media if thiamine was supplemented to the medium, making it an ideal biosensor to detect thiamine. Co-culturing these organisms together enabled Cryptococcus to grow to ~100-fold greater cell number than in pure culture, indicating that Variovorax was likely producing this vitamin. To confirm this, we then assayed the media during the growth of Variovorax and found it had produced ~100 mg/L of thiamine (Figure 4), far more than typical thiamine-producing bacteria (0.5-10 mg/L).
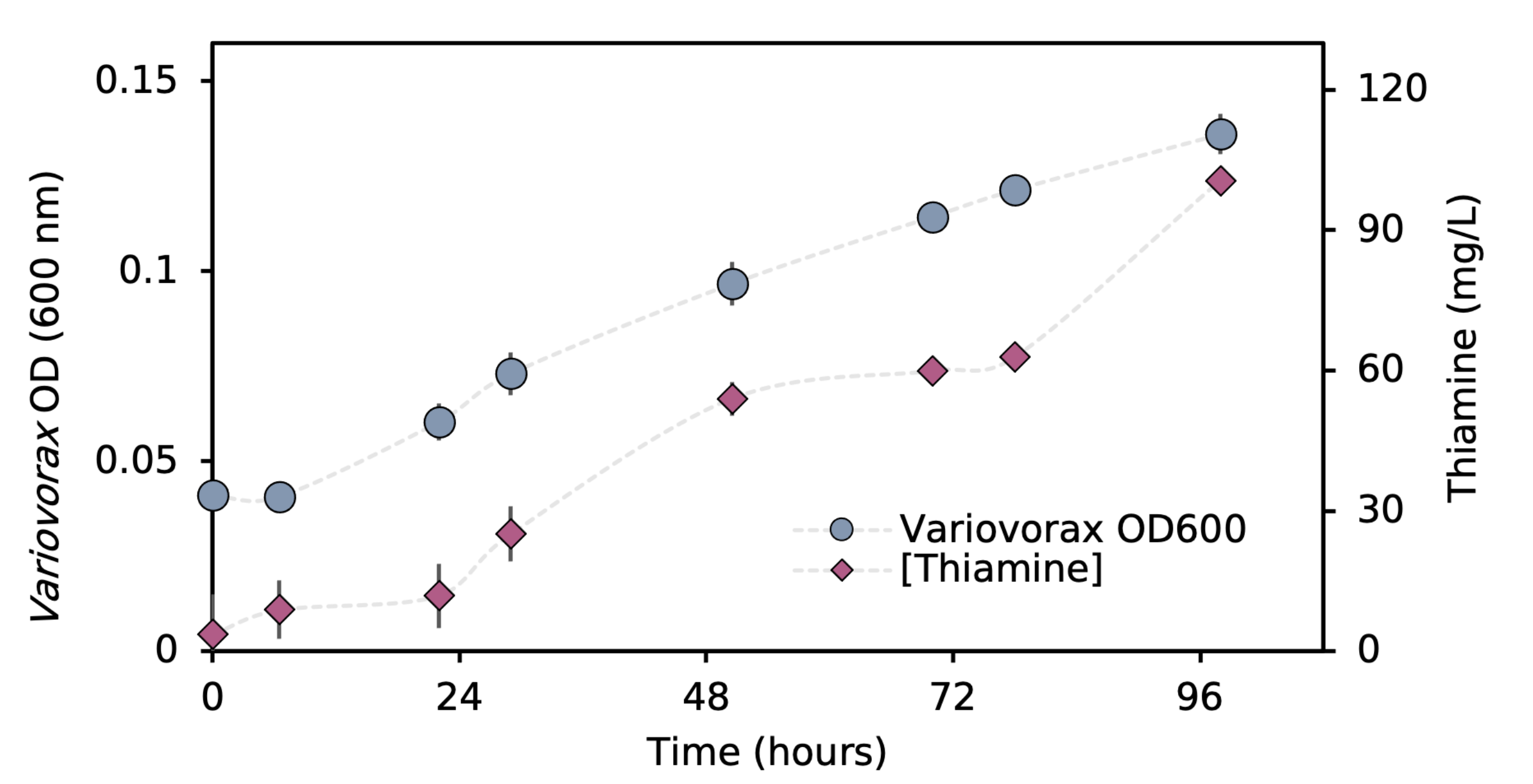
Interestingly, we found Variovorax was unable to produce its own pantothenate (vitamin B5), and supplying this vitamin to Variovorax shortens its lag phase and increases the amount of thiamine it produces. This led us to hypothesize that this organism's almost extreme thiamine-producing trait has evolved to encourage the growth of other bacteria that provide Variovorax with other compounds including pantothenate. The study demonstrates the value of combining metagenomics and network analyses to detect microbial interactions and, importantly, determine their mechanisms. This enables us to gain a better understanding of the metabolic functioning of particular organisms and microbial communities as a whole.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in