Consensus is key: reliable cryo-EM particle picking leveraging multiple pickers using REPIC
Published in Cell & Molecular Biology and Computational Sciences
Cryogenic electron microscopy (cryo-EM) has become a powerful technique for the structural determination of biological macromolecules. Despite its advancements, the early image processing step of particle picking—identifying individual macromolecular complexes within noisy micrographs—remains a significant bottleneck. This step is challenging due to low signal-to-noise ratios (SNR) of cryo-EM micrographs and lack of ground truth for particle locations, often requiring laborious manual intervention to identify useful particle sets.
Cryo-EM researchers rely on computational algorithms (blob picking, template matching, machine learning, etc.), known as particle 'pickers', to automate particle picking. However, pickers are inconsistent, often selecting different particle sets from the same micrograph. Picker inconsistency complicates the process of determining the best picker for a specific protein. Furthermore, extensive manual effort is required to curate particle sets (i.e., removing false positives) and build models using these curated sets and algorithms.
Introducing REPIC
To address these challenges, we developed REliable PIcking by Consensus (REPIC), an ensemble learning framework for cryo-EM particle picking. REPIC employs multiple pickers to identify a consensus set of high-quality particles, increasing the accuracy of this early step of cryo-EM image processing. The core principle of REPIC is that particles consistently identified by multiple pickers are more likely to be true positives, which allows REPIC to produce reliable particle sets for downstream image processing.
REPIC frames consensus particle picking as a graph theory problem, which is solved using integer linear programming (ILP). ILP allows for a globally optimal solution to be found that maximizes both the overlap of picked particles and their confidence scores from various pickers.
How REPIC Works:
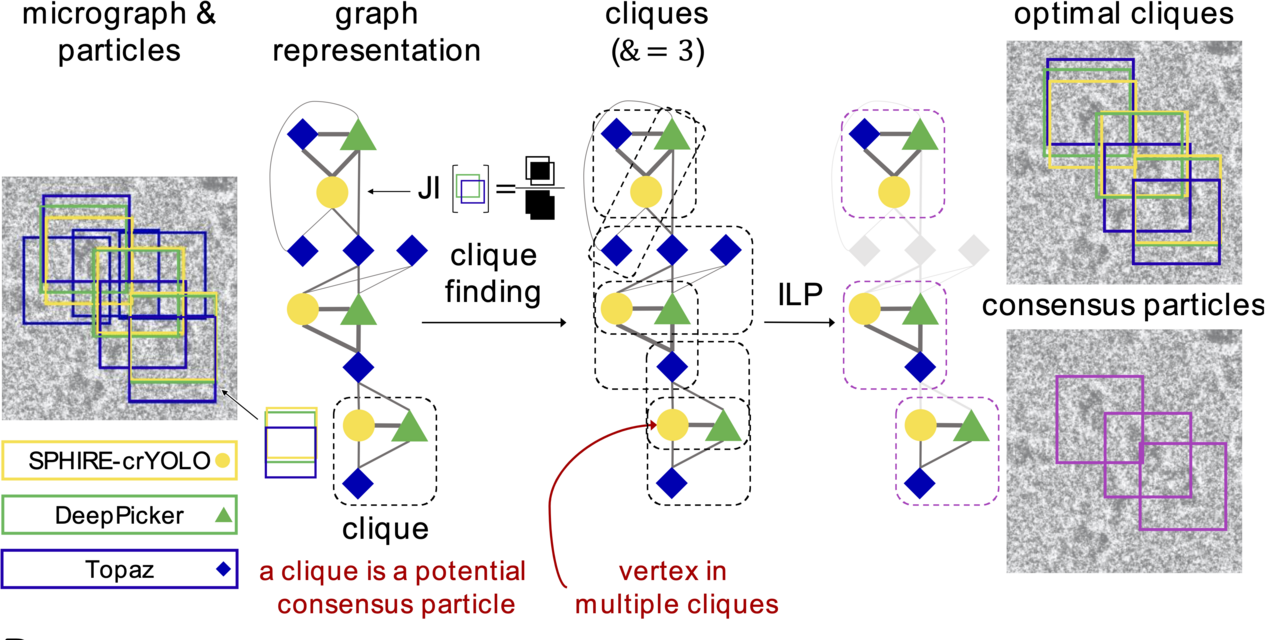
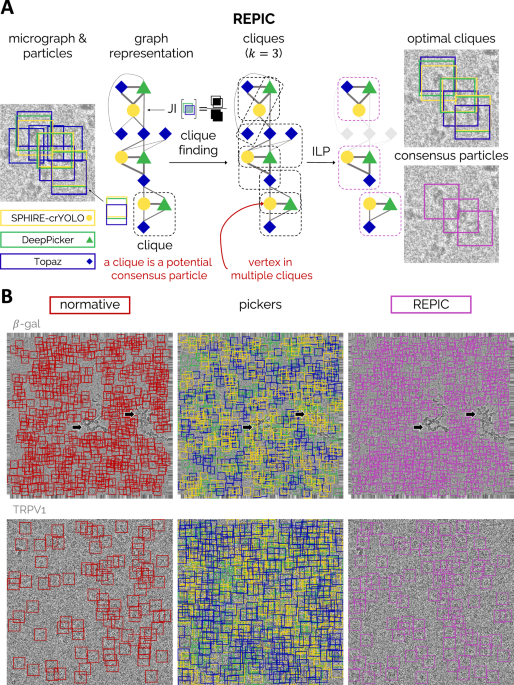
- Graph Building: Each particle identified by a picker is represented as a vertex in a computational graph. Edges are created between vertices based on the bounding box overlap, measured by the Jaccard Index. Edge weights correspond to the extent of the overlap, while vertex weights represent the confidence score from the picker.
- Clique Finding: REPIC then identifies cliques of size k in the graph, where k corresponds to the number of pickers. A clique indicates a subset of bounding boxes from various pickers that share substantial overlap and represents a potential consensus particle.
- Clique Optimization: Not all identified cliques are equally reliable. Some may exhibit high overlap but low confidence scores. REPIC uses ILP to select an optimal subset of cliques, maximizing both overlap and individual confidence scores. This ensures that the final particle set is composed of the most reliable consensus particles.
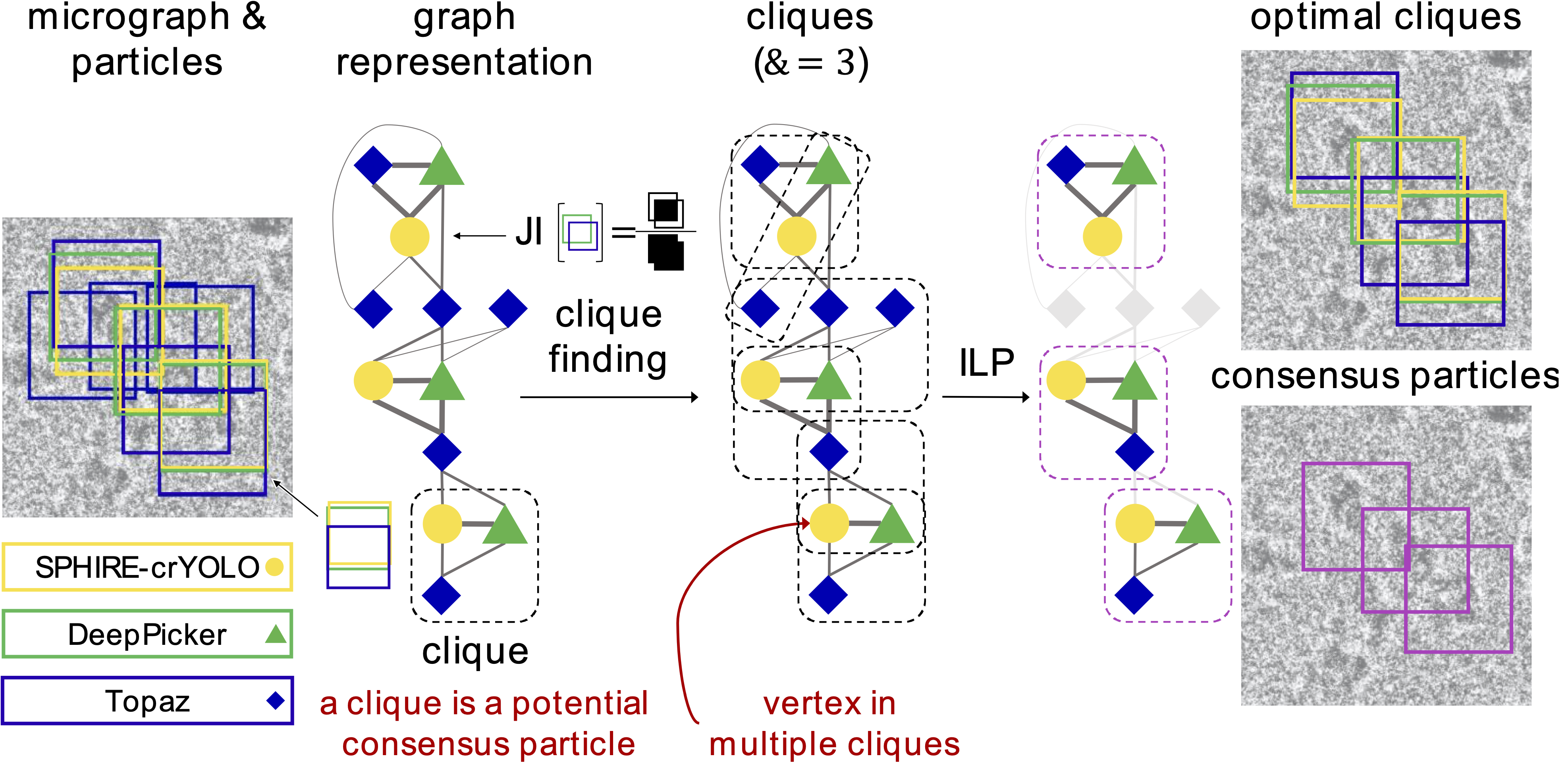
REPIC’s combination of graph theory and ILP optimization (Figure 1) produces a consensus set that represents the most consistent and reliable particles across different pickers.
Modes of Operation
REPIC has two modes to accommodate user needs:
- One-Shot Mode: This mode is useful when pre-trained pickers are expected to perform adequately. It streamlines the process of identifying high-quality particles without requiring additional picker training or choosing a specific picker.
- Iterative Mode: The iterative mode is designed for new datasets where pre-trained pickers underperform, or initial training examples are limited. By retraining the pickers using consensus sets from previous iterations, REPIC improves individual picker performance. This iterative process improves both precision and recall, allowing users to start with either minimal manually picked particles or one-shot outputs.
Performance and Benefits of REPIC
REPIC was evaluated using six cryo-EM datasets (EMPIAR-10005, 10017, 10057, 10093, and 10454, 12287) and three CNN-based pickers—SPHIRE-crYOLO 1, DeepPicker 2, and Topaz 3:
- Robustness: REPIC effectively identifies high-quality consensus particle sets even when individual pickers perform poorly. This reliability makes it a valuable tool, especially when the optimal picker is unknown.
- Reduced Manual Intervention: By significantly decreasing the need for manual picking, REPIC saves researchers time and effort, particularly in its iterative mode, which can yield high-resolution 3D maps from minimal initial training examples.
- Improved Picker Performance: Iterative applications of REPIC improved the performance of all tested pickers, aligning results towards more consistent particle sets. This indicates that REPIC effectively enhances existing pickers, especially with limited training data.
- Handling Heterogeneity: REPIC identifies consensus particles even in heterogeneous datasets, as demonstrated by its performance with the NOMPC dataset (EMPIAR-10093). This capability broadens its application to complex biological systems where heterogeneity is present.
Addressing Limitations
Despite its advantages, REPIC has limitations:
- Reliance on Existing Pickers: The effectiveness of REPIC hinges on the quality of the employed pickers. If all pickers underperform, the consensus approach will fail, emphasizing the need to choose appropriate pickers and initial training examples.
- Computational Demands: The most expensive step in REPIC's iterative mode is the retraining of individual pickers.
Conclusion
REPIC advances cryo-EM particle picking by integrating the strengths of multiple particle pickers into an ensemble, reducing manual intervention, and producing 3D maps comparable to those obtained from expert-picked particle sets.
For more information about REPIC, please refer to the published manuscript in Nature Communications Biology: https://www.nature.com/articles/s42003-024-07045-0
Software availability
REPIC is made free and open source on GitHub: https://github.com/ccameron/REPIC
References
- Thorsten Wagner, Felipe Merino, Markus Stabrin, Toshio Moriya, Claudia Antoni, Amir Apelbaum, Philine Hagel, Oleg Sitsel, Tobias Raisch, Daniel Prumbaum, Dennis Quentin, Daniel Roderer, Sebastian Tacke, Birte Siebolds, Evelyn Schubert, Tanvir R. Shaikh, Pascal Lill, Christos Gatsogiannis, and Stefan Raunser. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Communications Biology, 2(1), June 2019. doi: 10.1038/s42003-019-0437-z.
- Feng Wang, Huichao Gong, Gaochao Liu, Meijing Li, Chuangye Yan, Tian Xia, Xueming Li, and Jianyang Zeng. DeepPicker: A deep learning approach for fully automated particle picking in cryo-EM. Journal of Structural Biology, 195(3):325–336, September 2016. doi: 10.1016/j.jsb.2016.07.006.
- Tristan Bepler, Andrew Morin, Micah Rapp, Julia Brasch, Lawrence Shapiro, Alex J. Noble, and Bonnie Berger. Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nature Methods, 16(11):1153–1160, October 2019. doi: 10.1038/s41592-019-0575-8.
Follow the Topic
-
Communications Biology

An open access journal from Nature Portfolio publishing high-quality research, reviews and commentary in all areas of the biological sciences, representing significant advances and bringing new biological insight to a specialized area of research.
Related Collections
With Collections, you can get published faster and increase your visibility.
Signalling Pathways of Innate Immunity
Publishing Model: Hybrid
Deadline: Feb 28, 2026
Forces in Cell Biology
Publishing Model: Open Access
Deadline: Apr 30, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in