Corals practice social distancing to avoid infection
Published in Microbiology

Explore the Research

Microscale tracking of coral-vibrio interactions - ISME Communications
ISME Communications - Microscale tracking of coral-vibrio interactions
The ongoing deterioration of coral reefs over the past decades has brought into focus the interactions between reef-building corals and their bacterial pathogens. To shed a new light on this problem we have taken a microscopy-based approach in order to directly visualize the onset of coral disease at spatial and temporal scales relevant to the pathogens.
The project began as an international HFSP-funded collaboration between Prof. Assaf Vardi at the Weizmann Institute of Science (Israel), Prof. Roman Stocker, then at MIT (USA), and Prof. Justin Seymour at University of Technology Sydney (Australia). A central goal of the project was to bring advanced microscopy and microfluidics to the study of coral disease. We chose a well-established system, the Indo-Pacific coral Pocillopora damicornis and its bacterial pathogen Vibrio coralliilyticus. Based on previous studies we assumed that chemotaxis might be involved in host recognition and in the establishment of infection. Indeed, a series of experiments involving the tracking of individual Vibrio cells swimming in constructed gradients of coral mucus demonstrated that V. coralliilyticus is attracted to the mucus, and more specifically to coral-excreted DMSP, a known chemo-attractant in the marine environment (Garren et al. 2014 ISME J.). This made sense, as the general assumption in the literature was that corals exchanged dissolved gasses and nutrients with their environment by simple diffusion. Thus bacteria could locate a coral simply by swimming up a gradient of whatever diffuses from the coral surface.
The obvious next step was to try and repeat these experiments with actual corals. But how do you get a living coral under a microscope? We began with a straightforward approach, breaking the coral into small fragments and picking the tiny tissue pieces, half a millimeter or less, that slough off. Placing these in a microfluidic chamber, together with some free swimming Vibrios, we expected to see the bacteria accumulating around the coral, perhaps even settling on it. To our surprise, the bit of coral tissue appeared to be generating its own flow, apparently rapid enough to sweep away any approaching pathogens (Figure 1A). Further study confirmed that the coral surface was completely covered in beating cilia (Figure 1B), producing currents 10 times as fast as a swimming Vibrio (Shapiro et al. 2016, PNAS). Naturally, this result completely changed our perception of the coral surface micro-environment, and of how invading pathogens may be interacting with it.
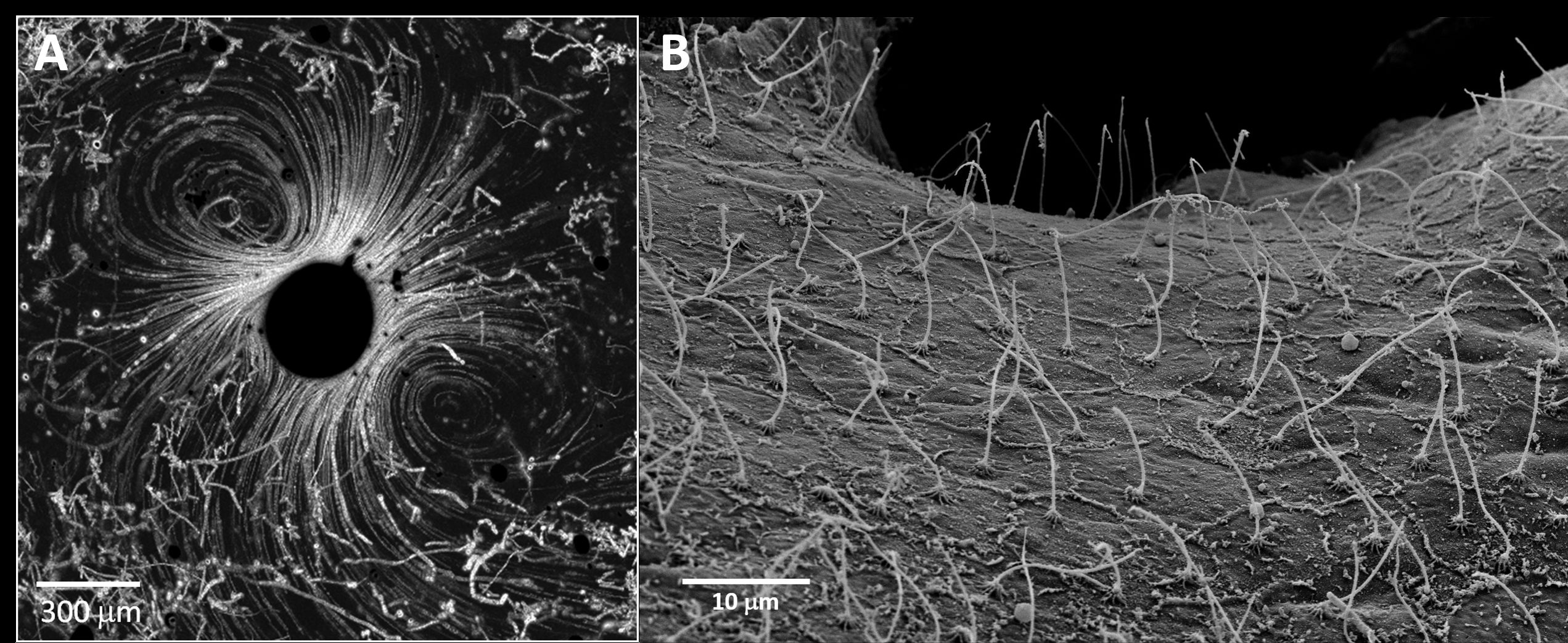
Figure 1. Corals use cilia to constantly stir the water near their surface. A. A small piece of P. damicornis tissue (dark circle in the center of the picture) surrounded by swimming V. coralliilyticus cells. The strong ciliary flows at the tissue surface, 10 times the swimming speed of the pathogen, effectively prevent bacteria from settling on the coral surface. B. Scanning electron micrograph showing a carpet of cilia covering the surface a P. damicornis coral.
We next wished to bring a living polyp, the basic unit of a coral colony, under the microscope, in the hope of finding the chink in the armor, where Vibrios could invade. After some effort we succeeded in establishing a protocol for inducing individual P. damicornis polyps to abandon the coral skeleton, via a process known as polyp bail-out, and to settle inside a microfluidic chamber pre-fabricated on a glass microscope slide (Shapiro et al 2016, Nat Comm). We were now able to keep the polyp under the microscope for hours, or even days. By adding fluorescently labeled Vibrios into the chamber we were able to see where they went. Search as we may, we could not find any labeled Vibrios settling on the polyp surface. However, looking through the glass slide we could see multiple Vibrio cells inside the polyp gut. So was it an upset stomach that killed the coral? We couldn’t say for sure. The settled polyps left the glass surface soon after the onset of infection, and we never got to see the disease progress further.
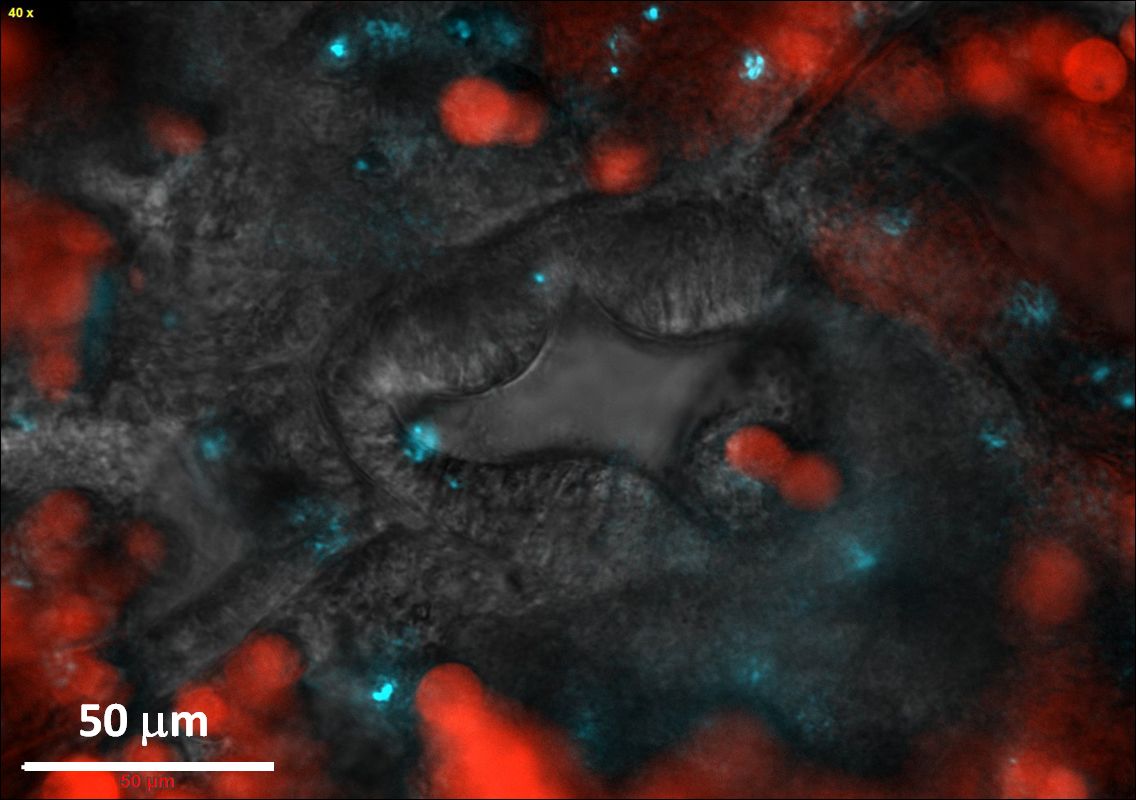
Figure 2. Vibrio pathogens rapidly accumulate in the coral gut. The unique view afforded by the "Coral on a Chip" system enabled us to capture this image just minutes after inoculation of the system with V. coralliilyticus. The polyp pharynx, seen here from the inside, is surrounded by symbiotic algae (red) and fluorescently labeled pathogens (cyan) (image taken from Shapiro et al., 2016, Nat com.)
But it was the ones that got sick that we were really interested in. Starting about 2 h from inoculation, the polyps appeared to be practicing social distancing, the tissue connecting them stretching and ripping. Rather than a colony, the coral now became a group of isolated polyps separated by bare skeleton. To our surprise, many of these polyps managed to survive the infection even while their immediate neighbors succumbed to it, their natural GFP signal decaying and their tissue visibly disintegrating. While a few of these polyps remained attached to the skeleton, the majority opted for bailing-out, detaching from the coral skeleton to become a free polyp. We were even able to retrieve these polyps, some of which remained viable for weeks following the infection. In nature, this behavior could perhaps facilitate the resettlement of such polyps, to start a new colony on some suitable location. On just a few occasions we saw another mode of infection. Tiny lesions in the coral tissue, perhaps inflicted while transferring them to the chambers, were rapidly colonized by labeled Vibrios. Perhaps here, where ciliary flow was disrupted, Vibrios were able to chemotax towards the coral and settle on its surface.
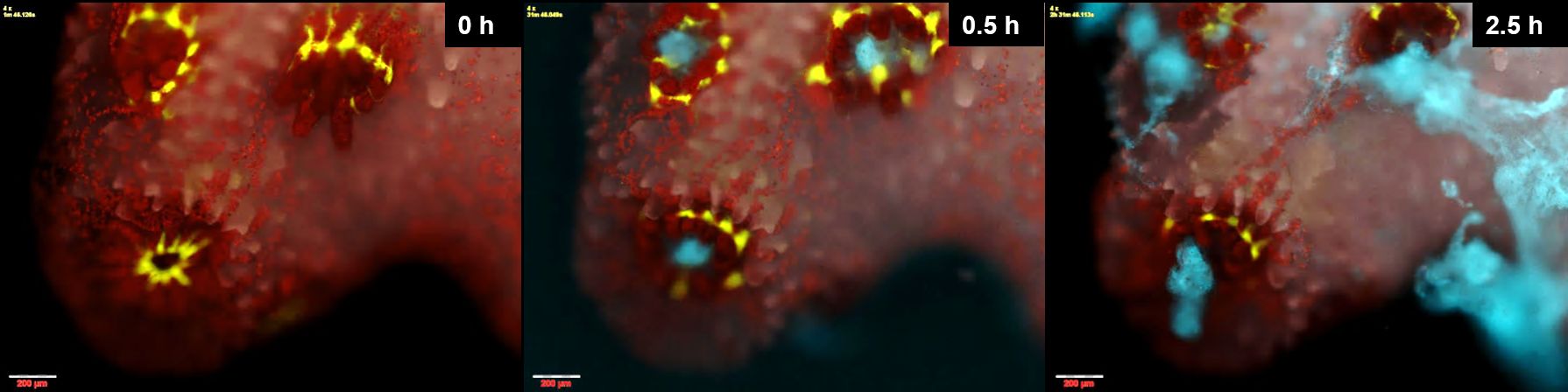
Figure 3. Coral polyps practicing social distancing to avoid infection. A small P. damicornis colony held in a microfluidic chamber is infected by V. coralliilyticus. Polyps are seen surrounded by a ring of green fluorescent protein, with symbiotic algae seen in red. Within half an hour of inoculation, fluorescently labeled pathogens (cyan) accumulate in the polyps' pharynx. Two hours later the polyps are separated, and plumes of pathogen-laden mucus are spewed from their guts, possibly in an attempt to rid themselves of invading pathogens (image taken from Gavish et al., 2021, ISME com.)
Further insights were gained from analyzing the water collected at the exit of the infection chambers. We specifically analyzed the activity of metalloproteases (MMPs), previously suggested to play a key role in the virulence of V. coralliilyticus. We saw a large peak in activity coinciding with the 2h inoculation period. Another peak was consistently observed towards the end of the infection, after the coral was apparently dead. This peak was invariably followed by a peak in bacterial abundance in the outflowing water. This sequence suggested an alternative role for the MMPs, that of breaking down the dead coral tissue to facilitate the rapid proliferation of the pathogen, and its subsequent release to the surrounding water.
The results of this quest gave us several new insights into the microscale landscapes of coral surfaces, and how coral pathogens may navigate them. This most recent work demonstrates that that V. coralliilyticus may colonize and infect open wounds, but that its primary mode of infection appears to be through the gut. In that it is surprisingly similar to the way Vibrios infect other hosts, including humans. We do not yet understand how the Vibrio enters the gut. Is it just through the intake of water by the coral polyp, or is the pathogen's own motility somehow involved? The surprisingly high dose required to initiate an infection suggests some means of delivery of high Vibrio concentrations to the coral. Could Vibrios reach the gut as coral ingest infected zooplankton, making coral disease a form of food poisoning? Other open questions concern the precise cues that inform the coral of a Vibrio infection, causing it to put up its guard. The role of other members of the coral microbiome in the infection process is also of interest. While some may help defending their host, others seem to benefit from the sudden surge of nutrients released by the infection. These and many other open questions we hope may be answered through future use of experimental systems similar to the ones presented here, bringing us a step closer to understanding, and possibly mitigating, coral disease and the demise of coral reefs.
References:
Garren, M., Son, K., Raina, JB. et al. ISME J 8, 999–1007 (2014). https://doi.org/10.1038/ismej.2013.210
Shapiro, O.H., Fernandez, V.I., Garren M., et al. Proceedings of the National Academy of Sciences 111(37), 13391-13396 (2014).
https://doi.org/10.1073/pnas.1323094111
Shapiro, O., Kramarsky-Winter, E., Gavish, A. et al. Nat Commun 7, 10860 (2016). https://doi.org/10.1038/ncomms10860
Gavish, A.R., Shapiro, O.H., Kramarsky-Winter, E. et al. ISME Commun 1, 18 (2021). https://doi.org/10.1038/s43705-021-00016-0


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in