Culturomics-based metagenomics (CBM) - a powerful approach for mining microbial dark matter resources hidden in unexplored habitats
Published in Microbiology

Poster image story: The roots of a Calligonum leucocladum in the desert extend hungrily for dozens of metres in all directions. Although its main trunk standing on the ground has dried up without any vitality, a tiny shoot is growing on the root in hope ...
Deserts are among the largest and most understudied biomes on Earth, covering about one-third of the total global land surface. Despite the general awareness of extremophilic microorganisms, deserts are generally regarded as lifeless and inhospitable ecosystems. In the past almost 20 years, an amazing microbial diversity and a huge biotechnological potential were unraveled using molecular approaches. Many related studies have revealed that deserts are an important reservoir of Earth’s microbial diversity, such as those spotlighting the Atacama Desert and polar deserts. However, due to strong niche specialization, difficulty in sampling, and limited adaptability of conventional culture methods, most of the desert microbes have been neglected to be cultivated and characterized in the laboratory. This large and poorly explored portion is colloquially called “Microbial Dark Matter (MDM)”, which represents a fundamental impediment to microbial ecology and bioresource exploitation.
Microbiome research based on direct metagenomic sequencing (e.g., 16S amplicon or shotgun sequencing) or culturomics continues to make exciting and important findings, but there are still some limitations with each of these two approaches. For example, the utility of direct metagenomic sequencing data is highly dependent on the complexity and biomass of the community, the sequencing method, and the public reference database, which may result in large numbers of low-abundance bacterial taxa or specific taxa being overlooked in the community. On the other hand, culturomics is generally considered to be labor-intensive and may miss important specific taxa in the community, therefore, making it difficult to obtain a comprehensive dataset of bacterial strains. It has been suggested that selective culture enrichment to reduce community complexity may be useful for metagenomic studies in some specific environments. However, research combining both culturomics and culture-enriched metagenomic sequencing approaches to mine desert microbiomes has not yet been carried out.
Based on this knowledge, we hypothesize that culture-enriched metagenomic sequencing based on a large-scale culturomic approach and high-resolution analysis enables us to fill the missing parts of the soil microbiome in direct sequencing.
Setting out to evaluate the effectiveness and prospect of the multi-omics strategy in desert soil microbiome research, we took the mining of microbial dark matter in desert soils as an example here, and present an integrated strategy that merges culturomic and metagenomic approaches (full-length 16S amplicon and shotgun sequencing), i.e., culturomics-based metagenomics (CBM) (Fig. 1). A total of 60 carefully selected or designed media, three incubation temperature (15, 30, and 45℃) were employed for each soil sample (including bulk and rhizosphere soils) to conduct the culturomics, resulting in 1800 agar plates. After the cultivation phase, the biomass was collected, DNA was extracted, and subjected to full-length 16S rRNA amplicon and shotgun sequencing. Meanwhile, we also conducted a second round of isolation on some frozen culture stocks to evaluate the effectiveness of the CBM-guided isolation strategy.
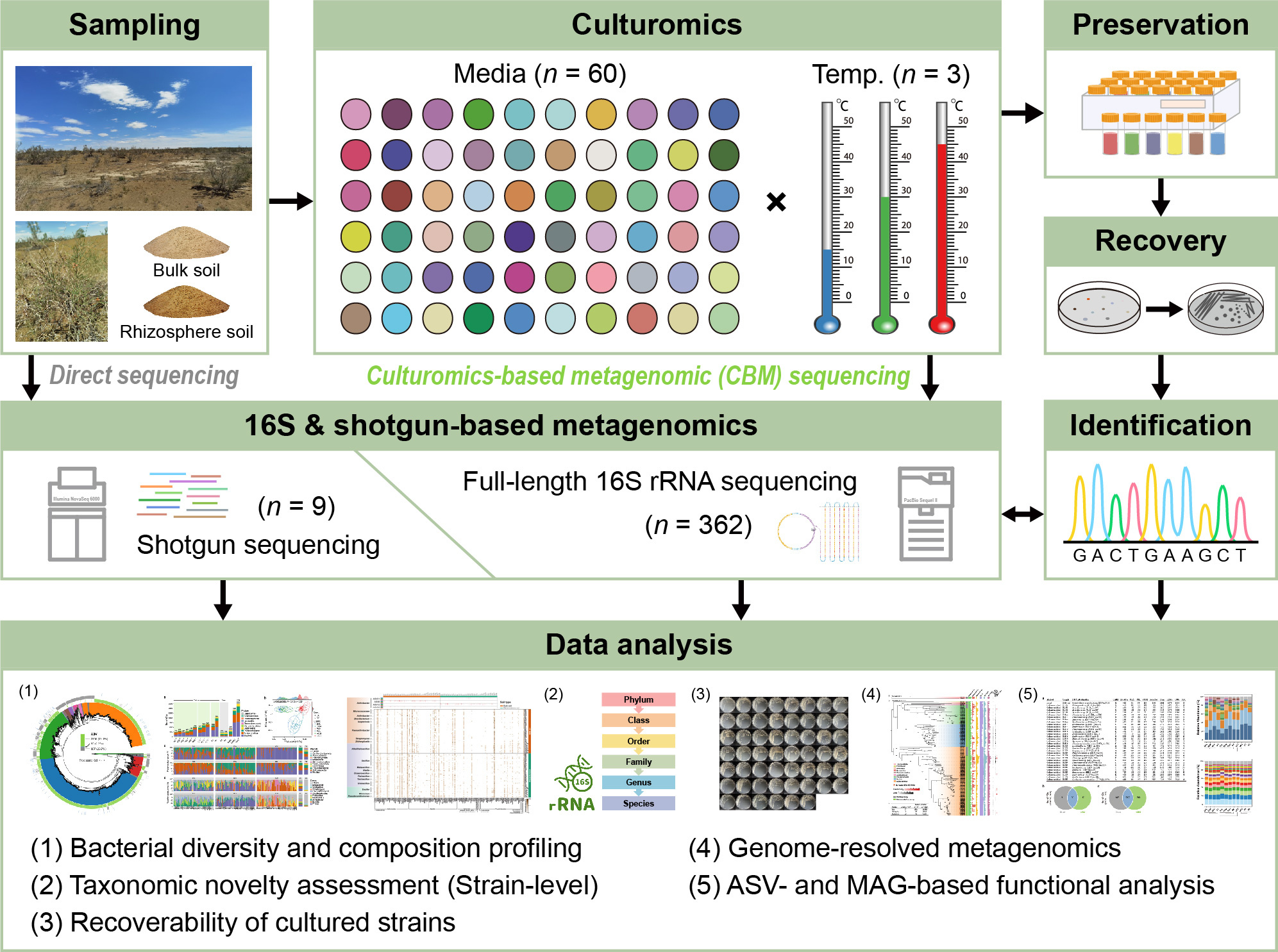
Fig. 1. Schematic diagram of the experimental design
In this work, we found that the integration of the high-resolution CBM strategy with direct metagenomic sequencing enables in-depth profiling of the microbial dark matter in desert soils. We demonstrated that CBM is a ideal strategy that can greatly improve the taxonomic and functional resolution of desert soil microbiome, and importantly it allows the post hoc recovery of microbes of interest based on the metagenomics-guided isolation. By species-level analysis, we also revealed the huge underexplored potential of novel bacterial resources in desert soils. Furthermore, the results of culturomics under multiple conditions provide an important reference for the isolation of certain special or novel bacterial taxa in desert soils. With these data in hand, we can better understand the composition and distribution, microbe-microbe interactions, environmental adaptation mechanism, and gene repertoire of desert microbiota. Culturomics-based metagenomics, as exemplified strategy here for desert soils, provides a new perspective for deeper understanding and mining microbial dark matter in microbiome samples, especially those from extreme habitats.
Follow the Topic
-
npj Biofilms and Microbiomes

The aim of this journal is to serve as a comprehensive platform to promote biofilms and microbiomes research across a wide spectrum of scientific disciplines.
Related Collections
With Collections, you can get published faster and increase your visibility.
Natural bioactives, Gut microbiome, and human metabolism
Publishing Model: Open Access
Deadline: Feb 20, 2026
Harnessing plant microbiomes to improve performance and mechanistic understanding
Publishing Model: Open Access
Deadline: Jun 01, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in