Deep learning large-scale drug discovery and repurposing
Published in Bioengineering & Biotechnology, Chemistry, and Computational Sciences
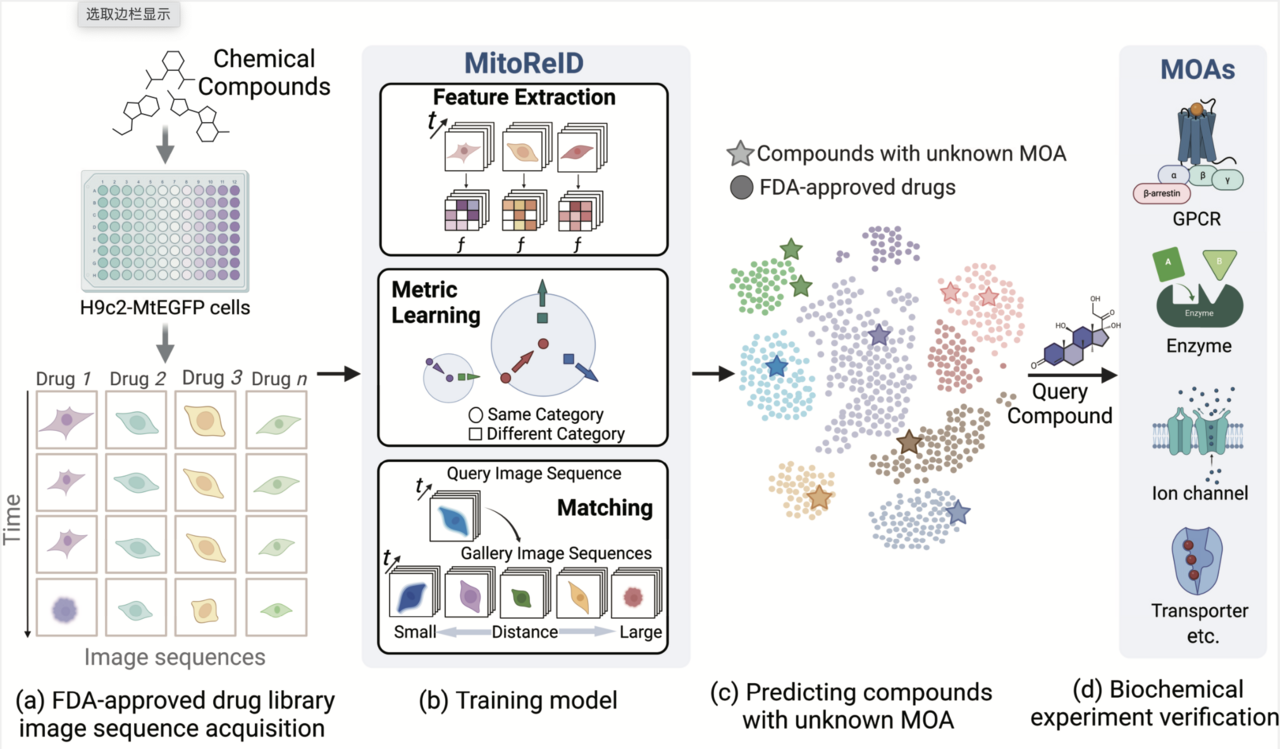
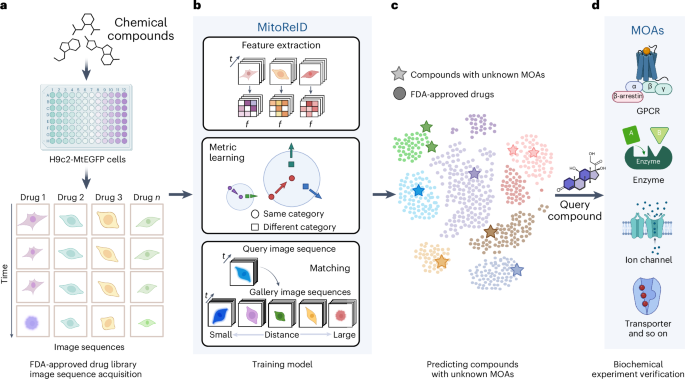
Large-scale drug discovery and repurposing is challenging. Identifying the mechanism of action (MOA) is crucial, yet current approaches are costly and low-throughput. Here we present an approach for MOA identification by profiling changes in mitochondrial phenotypes. By temporally imaging mitochondrial morphology and membrane potential, we established a pipeline for monitoring time-resolved mitochondrial images, resulting in a dataset comprising 570,096 single-cell images of cells exposed to 1,068 United States Food and Drug Administration-approved drugs. A deep learning model named MitoReID, using a re-identification (ReID) framework and an Inflated 3D ResNet backbone, was developed. It achieved 76.32% Rank-1 and 65.92% mean average precision on the testing set and successfully identified the MOAs for six untrained drugs on the basis of mitochondrial phenotype. Furthermore, MitoReID identified cyclooxygenase-2 inhibition as the MOA of the natural compound epicatechin in tea, which was successfully validated in vitro. Our approach thus provides an automated and cost-effective alternative for target identification that could accelerate large-scale drug discovery and repurposing.
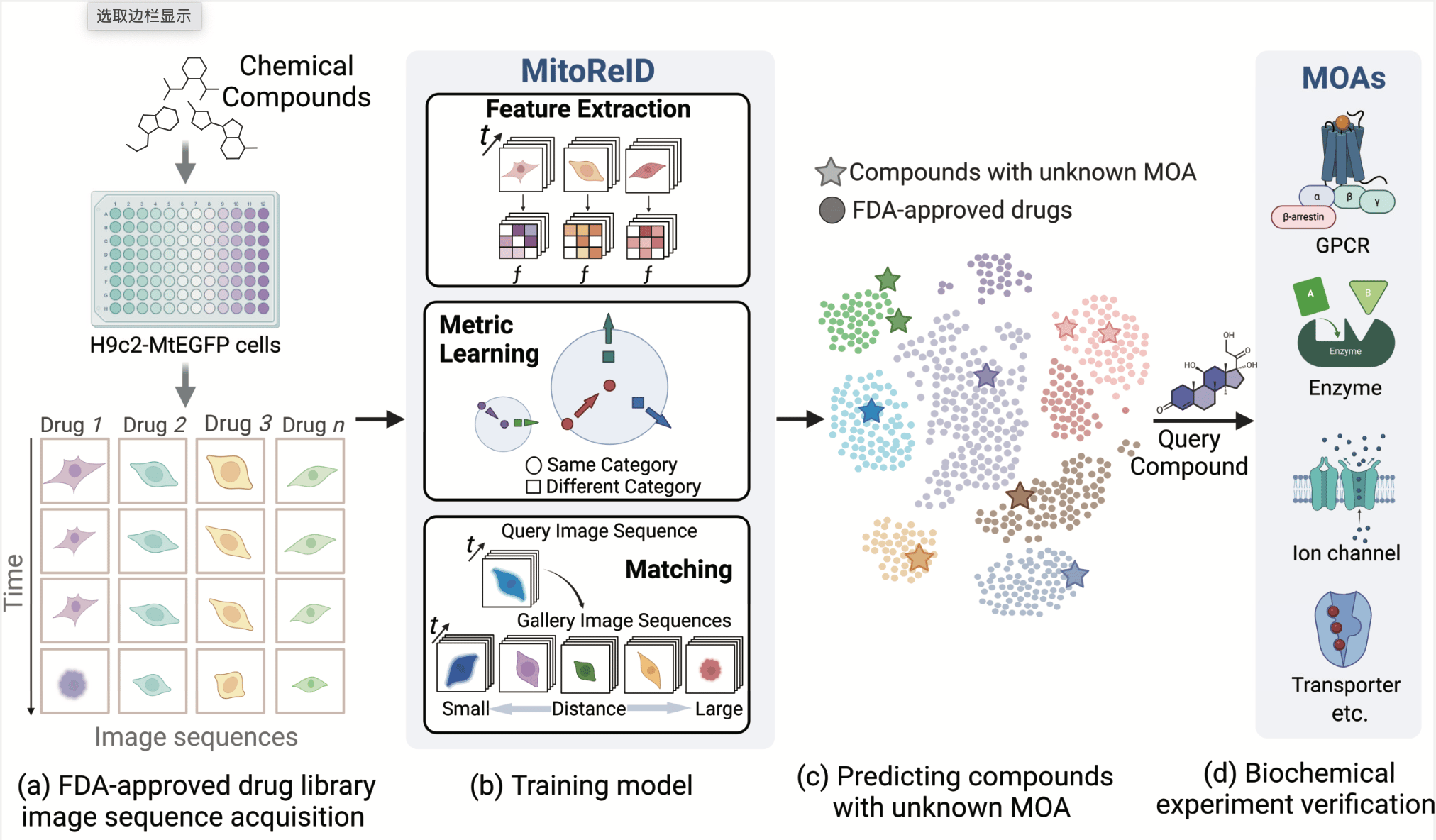
Fig. 1: Framework of deep learning-based MOA prediction by profiling temporal mitochondrial phenotype for large-scale drug discovery and repurposing.
-
Fig. 2: High-throughput acquisition of time-lapse images for temporal mitochondrial phenotypes. 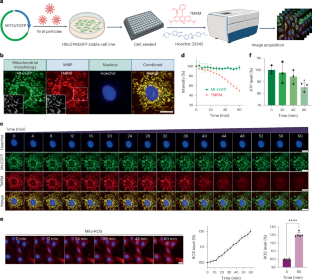
-
Fig. 3: Drugs with varied MOAs exhibit diverse mitochondrial phenotypes. 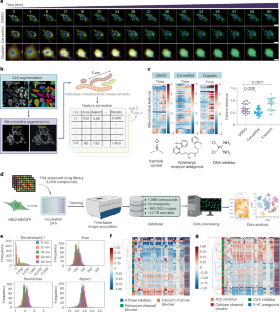
-
Fig. 4: Deep learning approaches for temporal mitochondrial phenotypes recognition. 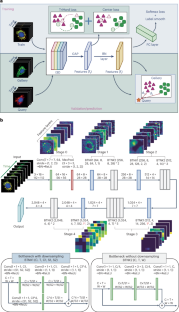
Follow the Topic
-
Nature Computational Science

A multidisciplinary journal that focuses on the development and use of computational techniques and mathematical models, as well as their application to address complex problems across a range of scientific disciplines.
Related Collections
With Collections, you can get published faster and increase your visibility.
Physics-Informed Machine Learning
Publishing Model: Hybrid
Deadline: May 31, 2026
Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in