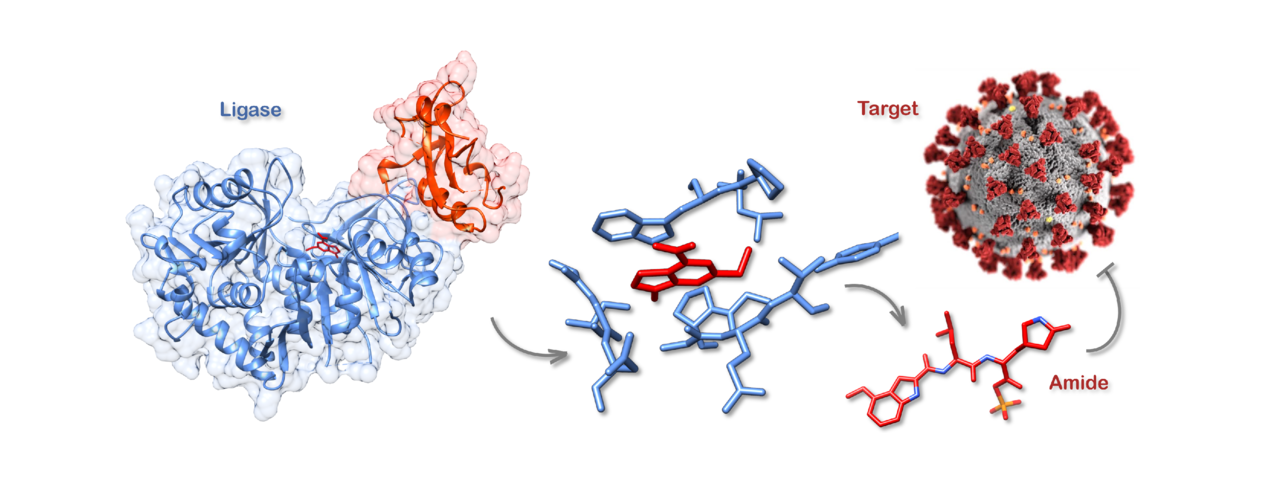
Amide bonds can be found throughout nature, they link peptides and proteins together and are also found in many naturally occurring bioactive molecules such as the antibiotics penicillin and vancomycin. In addition to being a fundamental reaction in nature, amide bond formation is also one of the most widely used transformations in synthetic chemistry. Synthesising an amide using traditional chemical techniques can be achieved by coupling a carboxylic acid and an amine together and is relatively simple to accomplish. However, it involves the use of deleterious chemicals, often requires protecting groups and typically requires multiple steps to install each amide bond. By-products formed during amide synthesis can also make purification of the final product laborious and time-consuming.
For many years, our laboratory has been interested in studying Streptomyces bacteria due to their ability to produce a variety of bioactive compounds with astounding structural diversity, many of which have important applications as pharmaceuticals. One of our bacterial strains, Streptomyces scabies, produces a compound called coronafacoyl-isoleucine (CFA-Ile), a member of the coronatine family of phytotoxins (compounds that are toxic to plants). Whilst studying the biosynthesis of phytotoxins in S. scabies, and related bacterial species, we discovered a family of adenosine triphosphate (ATP)-dependent ligase enzymes, called CfaL, that are responsible for the biosynthesis of CFA-Ile and related phytotoxins. CfaL couple the carboxylic acid CFA with amino acids such as L-isoleucine (L-Ile), to generate amides in a single step. Whilst other ligases have previously been explored for amide synthesis, these mainly produce thioester intermediates. A secondary acyl transferase enzyme is then required to make the final amide products. Since CfaL are able to couple acids and amines directly, without the need of a partner enzyme, we wondered if they could be applied more widely for the synthesis of compounds of interest containing amides.
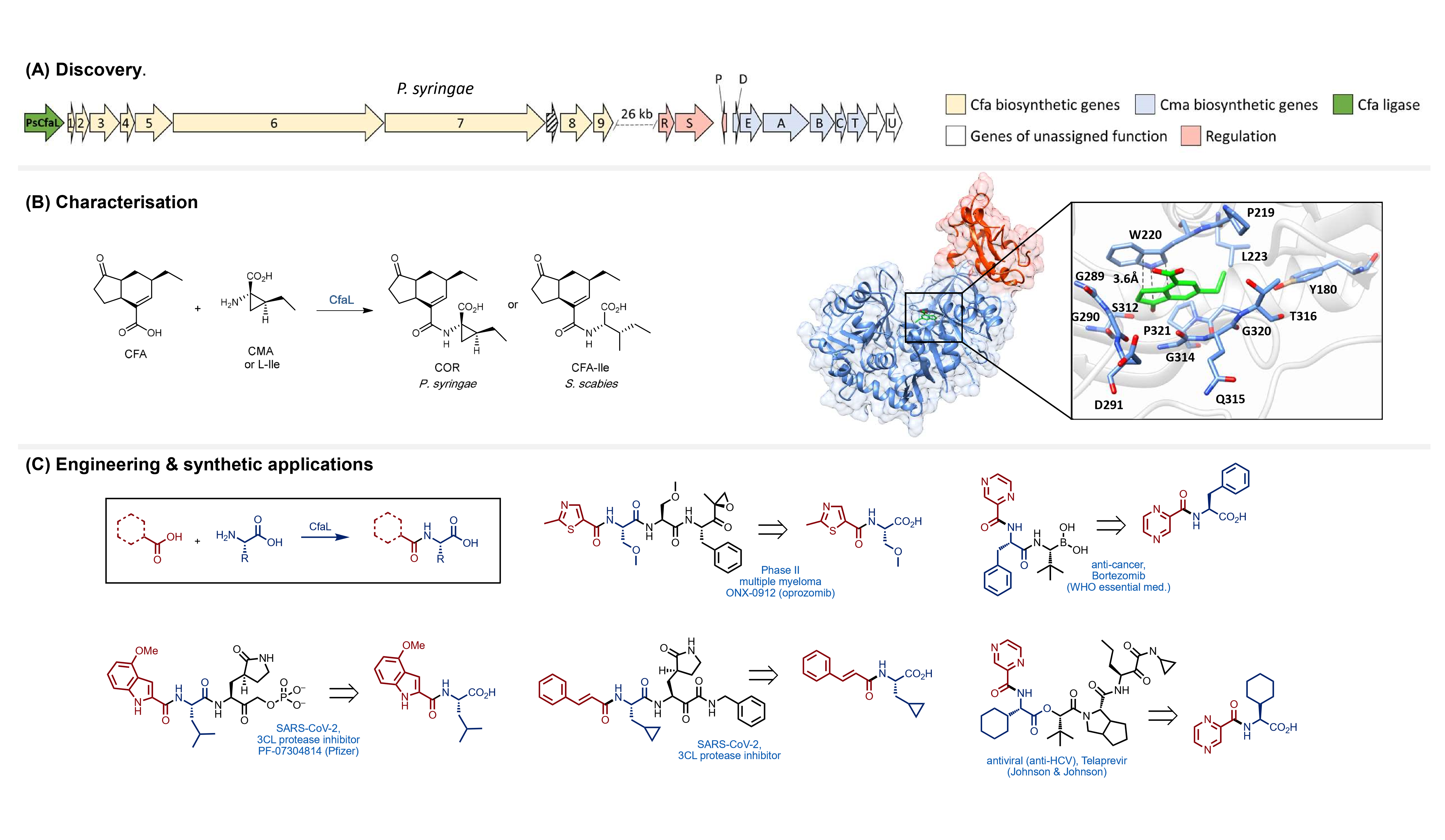
Figure 1: Discovery & development of ligases for amide synthesis: (A) CfaL enzymes were discovered by genome mining of bacteria producing coronatine-like phytotoxins; (B) CfaL enzymes were characterised to determine kinetic parameters, substrate scope and X-ray crystallography was used for structure determination; (C) Structure guided mutagenesis was then employed to engineer improved CfaL variants for drug synthesis.
CfaL were found to catalyse the synthesis of amides from a large variety of different carboxylic acid and amino acid substrates. The presence of additional reactive functional groups, in either the acid donor or amine acceptor substrates, did not interfere with the fidelity of enzymatic ligation reactions. For traditional coupling chemistry, reactive groups would typically require protecting, with each amide bond requiring three steps (protect-couple-deprotect) to construct. Members of the CfaL family can also be used for kinetic resolutions of racemic acid donor and amine acceptor substrates, for example enabling the selective amidation of the biologically active (S)-ibuprofen from a racemic mixture of two enantiomers. Solving the x-ray crystal structure of a CfaL enzyme provided valuable information which led to mutants, via structure guided mutagenesis, with improved stability and activities. Furthermore, we were able to demonstrate that these enzymes function efficiently at gram scale, producing excellent quantities of amide products that can be easily purified using a simple solvent extraction procedure. Some of the products we can make with CfaL have real word applications in the preparation of precursors for important pharmaceuticals such as cancer treatments or potential COVID-19 drugs. We believe that CfaL enzymes represent a new, more sustainable, alternative route for the production of amide containing pharmaceuticals, agrochemicals and other valuable materials.
This research has now been published in Nature.
https://doi.org/10.1038/s41586-021-03447-w
Michael Winn, Michael Rowlinson, Fanghua Wang, Luis Bering, Daniel Francis, Colin Levy & Jason Micklefield
Follow the Topic
-
Nature

A weekly international journal publishing the finest peer-reviewed research in all fields of science and technology on the basis of its originality, importance, interdisciplinary interest, timeliness, accessibility, elegance and surprising conclusions.


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in