Novel antibiotics are urgently needed to combat the growing problem of antimicrobial resistance, as well as to treat neglected diseases and possible future pandemics. Many antibiotics in the clinic today (e.g. penicillin, vancomycin and daptomycin) are microbial natural products assembled by enzymes called nonribosomal peptide synthetases (NRPS). These complex enzymes are comprised of a series of modules organised into an assembly line architecture, with each module introducing a single amino acid precursor into a growing peptide chain.
Their modular organisation suggests that manipulation of these NRPS could be relatively straightforward. In principle, replacement of modules within the NRPS assembly line should enable a different amino acid building block to be introduced at a selected position, leading to user-defined peptide products. In practice however, efforts to engineer NRPS enzymes have met with limited success. In most cases, the engineered enzyme variants have completely abolished activity or greatly reduced product yields. Moreover, previous NRPS engineering efforts have focused on relatively small NRPS enzymes (comprised of only a few modules) which are manipulated and produced in simple host species (e.g. E. coli). However, the majority of the most important nonribosomal peptides are produced by non-model organisms, particularly Streptomyces bacteria. The genomes of Streptomyces species are GC rich and the NRPS genes are also highly repetitive in sequence, making genetic manipulation via traditional methods highly challenging. We aimed to manipulate NRPS encoding genes directly in the native Streptomyces producer strain, as the native producer contains all of the unusual amino acids and other “ingredients” as well as the regulatory components necessary for production.
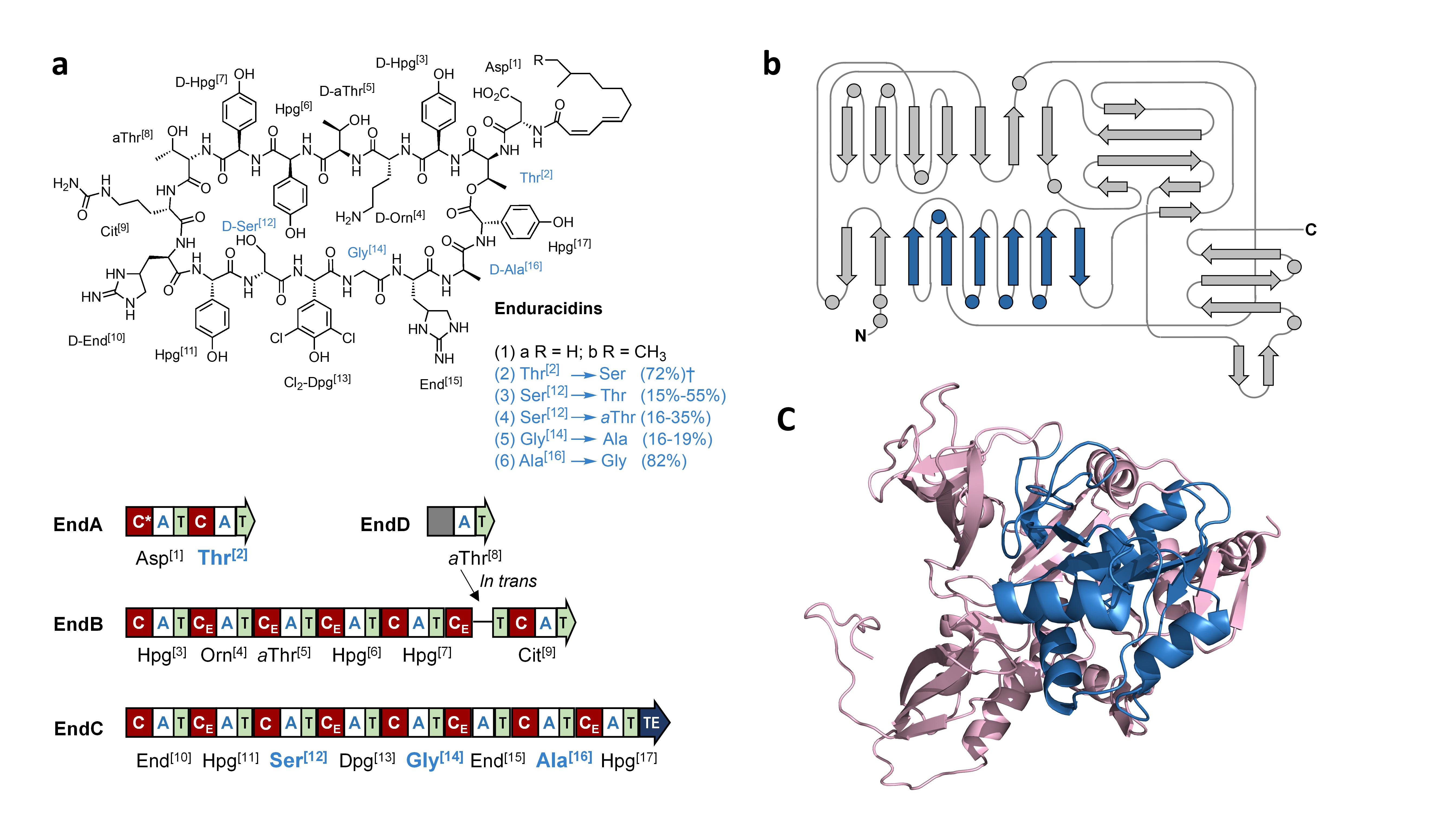
Figure 1: (a) structure of wild type and engineered enduracidins, amino acids in blue were targeted for modification, with organization of the enduracidin NRPS shown below; (b) secondary structure model with flavodoxin-like subdomain (FSD) shown in blue; (c) homology model of the Thr[2] A-domain, with the FSD shown in blue.
Our idea was to use CRISPR-Cas9 to edit the genes encoding for NRPS assembly lines in vivo, as this would enable precise replacement of key parts of the NRPS enzyme much more efficiently than using traditional methods. The increase in editing efficiency allowed us to manipulate the very large NRPS encoding genes involved in the biosynthesis of enduracidin in Streptomyces fungicidicus. Enduracidin is a lipopeptide antibiotic that is produced by an enormous NRPS consisting of 17 modules that assemble 17 amino acid precursors. Enduracidin is similar in structure and bioactivity to ramoplanin, which entered phase 3 clinical trials, and so it could be a potential lead-structure in developing new antibiotic treatments. By swapping out a small region within each module, a “subdomain” that recognises the amino acid substrates, we created new NRPS enzymes that produced modified peptide products with one amino acid building block swapped for another.
We found that the new CRISPR-Cas9 strategy was much more efficient and successful in delivering new lipopeptide antibiotics than the more traditional engineering approaches that had been used previously. Overall, we were able to produce 10 new lipopeptide variants by swapping subdomains that originated from a wide variety of bacterial species. Importantly, the new lipopeptides were produced in good yields relative to the wild type. Interestingly, we discovered that the sequence similarity of the subdomains being swapped was very important. Those subdomain swaps with higher similarity (i.e. those which were most closely related to the enduracidin subdomains) were much more likely to function within the engineered NRPS, and produce new lipopeptide products. We hope that this information will be a useful guide for future efforts using gene editing to engineer NRPS and possibly related assembly line enzymes such as polyketide synthases. We also performed whole genome sequencing on several of our edited strains, and saw that the CRISPR-Cas9 editing did not result in any major off-target effects, demonstrating how precise our method is. Finally, our method does not leave any scars or resistance genes (markers), and so it also lends itself to combinatorial editing where multiple subdomain swaps could be introduced into the same NRPS to further increase structural diversity of the assembly line products.
This research has now been published in Nature Communications.
https://doi.org/10.1038/s41467-021-27139-1
Wei Li Thong, Yinxin Zhang, Ying Zhuo, Katherine Robins, Joanna Fyans, Abigail Herbert, Brian Law & Jason Micklefield
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in