Divalent and multivalent cations control liquid-like assembly of poly(ADP-ribosyl)ated PARP1 into multimolecular associates in vitro
Published in Cell & Molecular Biology
Biomolecular condensation processes are studied intensively for the last 15 years, about 9000 articles have been published over the years. It is obvious that compartmentalization of cellular biomolecules into membraneless organelles or biomolecular condensates plays an important role in the regulation of biological processes. In particular, space- and time-specific formation of nuclear condensates is often associated with DNA repair. The formation of DNA repair compartments seems to be related with intensive synthesis of ADP-ribose polymer (poly(ADP-ribose), PAR) at DNA damage site events (Alemasova E. and Lavrik O., Nucleic. Acids Res., 2022).
We have been studied poly(ADP-ribose) polymerases (PARPs), namely PARP1 and PARP2, and their role in DNA repair for quite a long time. These nuclear enzymes are key cellular nick sensors and activated in a response to DNA damage. PARP1 and PARP2 use NAD+ as a substrate to synthesize the negatively charged polymer of ADP-ribose (PAR). Poly(ADP-ribose) is mainly covalently attached to PARP1 and PARP2, facilitating the dissociation of these enzymes from DNA damage site for subsequent repair of the damage. This was the classical hypothesis. However, many questions still remain to be addressed:
In collaboration with the laboratory of Pr. David Pastre (University of Evry – Val d'Essonne, France), we investigated the activation of PARP1 and PARP2 at DNA lesions using atomic force microscopy and observed the synthesis of PAR and PARP1 and PARP2 PARylation at the single-molecule level. We found that PARylated PARP1 and PARP2 molecules could persist at DNA lesions rather than dissociate, which led us to suggest that PAR covalently attached to PARP1(2) can initiate the formation of structures that recruit repair proteins to the lesion (Sukhanova M. et al, Nucleic. Acids Res., 2016). These experiments have already been carried out in the presence of Mg2+ cations that function as the cofactor for PARPs. In the presence of Mg2+ cations, we also observed the formation of supramolecular association of PARylated PARP1 and PARP2 using the dynamic light scattering method, which also indicated the formation of condensates (Vasil’eeva I. et al., Biochim. Biophys. Acta, 2019). Then, using AFM, we observed even large compartments formed by PARylated PARP1 and the RNA-binding protein FUS (Fused in Sarcoma) (Singatulina A. et al., Cell Rep., 2019). To our surprise, it was shown that damaged DNA is concentrated in these compartments.
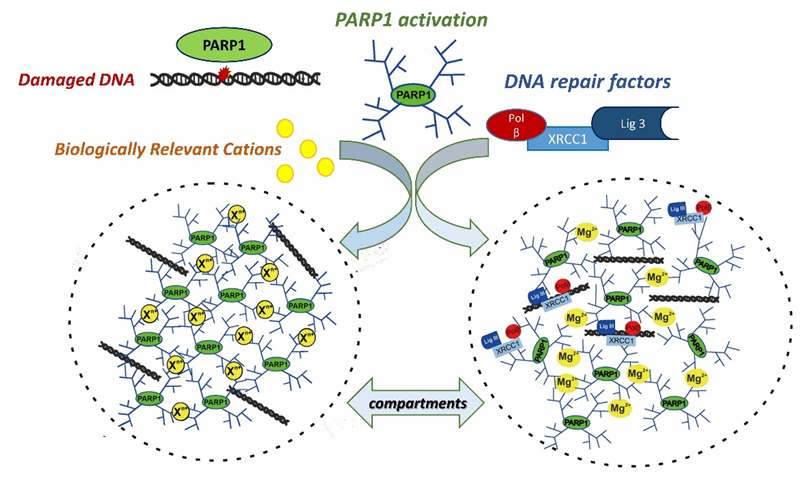
In the current paper, we were able to show that the formation of the compartments indeed occurs with the participation of PARylated PARP1 and repair proteins in the presence of bivalent metal cations or polyamines. One of the driving force to concentrate repair proteins in the compartments, might be their property to effectively interact with poly (ADP-ribose) which we showed earlier (Moor N. et al., 2020, Biochimie; Vasil’eva I. et al., 2021, Int. J. Mol. Sci.) and confirmed in this study.
Thus, data obtained in this paper show that the formation of repair condensate via liquid-like assembly of PARylated PARP1 is driven by biogenic cations. This assembly regulates PARP1 autoPARylation, hydrolysis of poly(ADP-ribose) catalyzed by poly(ADP-ribose) glycohydrolase (PARG) and DNA synthesis by DNA polymerase beta.
|
|
|
The proposed model of the cation-dependent liquid-like assembly of PARylated PARP1 and formation of repair condensates. |
Thus, this work explains the role of PAR and biology relevant cations in the formation of condensates and points to the direct involvement of PARylated PARP1 and PARP2 in the regulation of DNA repair via condensate formation. Therefore, PARP1 and PARP2 and poly(ADP-ribose) may serve as dynamic triggers in the formation of cellular compartments for DNA repair execution in a chromatin context, and the extension of this research to the cellular level should bring new discoveries about the regulation of DNA repair.
As PARP1 inhibition hold great promise in cancer therapy, PARP1-dependent condensates could represent a new class of drug targets that can be associated with the development of novel therapeutic approaches.
References
Alemasova, E. E., & Lavrik, O. I. A sePARate phase? Poly (ADP-ribose) versus RNA in the organization of biomolecular condensates. Nucleic Acids Res. 2022, 50, 10817-10838. DOI: 10.1093/nar/gkac866.
Sukhanova, M. V., Abrakhi, S., Joshi, V., Pastre, D., Kutuzov, M. M., Anarbaev, R. O., Curmi, P.A., Hamon, L. & Lavrik, O. I. Single molecule detection of PARP1 and PARP2 interaction with DNA strand breaks and their poly (ADP-ribosyl) ation using high-resolution AFM imaging. Nucleic acids Res. 2016, 44 e60-e60. DOI: 10.1093/nar/gkv1476.
Vasil'eva, I. A., Anarbaev, R. O., Moor, N. A., & Lavrik, O. I. Dynamic light scattering study of base excision DNA repair proteins and their complexes. Biochim Biophys Acta-Proteins and Proteomics. 2019, 1867, 297-305. DOI: 10.1016/j.bbapap.2018.10.009.
Singatulina, A. S., Hamon, L., Sukhanova, M. V., Desforges, B., Joshi, V., Bouhss, A., Lavrik, O.I. & Pastré, D. PARP-1 activation directs FUS to DNA damage sites to form PARG-reversible compartments enriched in damaged DNA. Cell Rep. 2019, 27, 1809-1821. DOI: 10.1016/j.celrep.2019.04.031.
Moor, N. A., Vasil’eva, I. A., Kuznetsov, N. A., & Lavrik, O. I. Human apurinic/apyrimidinic endonuclease 1 is modified in vitro by poly (ADP-ribose) polymerase 1 under control of the structure of damaged DNA. Biochimie. 2020, 168, 144-155. DOI: 10.1016/j.biochi.2019.10.011.
Vasil’eva, I., Moor, N., Anarbaev, R., Kutuzov, M., & Lavrik, O. (2021). Functional roles of PARP2 in assembling protein–protein complexes involved in base excision DNA repair. Int. J. Mol. Sci., 2021, 22, 4679. DOI: 10.3390/ijms22094679.
Follow the Topic
-
Communications Biology

An open access journal from Nature Portfolio publishing high-quality research, reviews and commentary in all areas of the biological sciences, representing significant advances and bringing new biological insight to a specialized area of research.
Related Collections
With Collections, you can get published faster and increase your visibility.
Signalling Pathways of Innate Immunity
Publishing Model: Hybrid
Deadline: Feb 28, 2026
Forces in Cell Biology
Publishing Model: Open Access
Deadline: Apr 30, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in