Duodenal bacterial proteolytic activity determines sensitivity to dietary antigen through PAR-2 receptor
Published in Microbiology

Here, we identify a bacterial enzyme, elastase, that causes immune activation and worsens reactions to gluten in genetically susceptible hosts.
Celiac disease is a life-altering food sensitivity triggered by ingesting gluten, the common protein found in wheat, barely, and rye. Gluten is used ubiquitously in the food industry, and is not only found in bread, pastas and baked goods, but is a common ingredient in processed sauces, salad dressings and can even be found in some medications. While the variety of gluten-free alternatives has vastly improved over the last decade, the complete avoidance of gluten is almost impossible- something those suffering from celiac disease must do to avoid symptoms and potentially severe complications. In order to find better ways of preventing or treating the disease, we need detailed understanding of the additional conditions required for development and progression.
The rapid increase in celiac disease prevalence suggests environmental factors are contributing to the disease-but how? Recent efforts have focused on the gut microbiota as a potential factor, given its role in determining health and disease. Indeed, clinical association studies have demonstrated shifts in microbial composition between individuals with celiac disease and those without celiac disease, but no "celiac microbial signature" has been defined. This is similar to what we have learned in other chronic inflammatory diseases, such as inflammatory bowel disease, and it may mean we should investigate collective functional signatures in addition to changes in microbial composition. To understand how microbes influence food sensitivity, we have focused on the proteolytic properties of microbes found in the small intestinal that could interact with pathways relevant to celiac disease pathogenesis.
As a part of his PhD, Alberto Caminero found that the small intestine harbours many bacterial groups that can degrade gluten, a group of proteins highly resistant to digestion by host enzymes in the small intestine. In the Verdu lab at the Farncombe Institute at McMaster University, Dr. Caminero further explored this concept as a post-doctoral fellow. He found that proteases from both opportunistic pathogens, like Pseudomonas aeruginosa, and commensal bacteria, like Lactobacillus, can participate in gluten metabolism but they generate different patterns of gluten peptides harbouring different immunogenic properties. However, proteases have diverse functions in the gut, and we hypothesized that proteolytic bacteria in the small intestine could influence other pathways involved in celiac disease pathogenesis, perhaps even independently from gluten.
In this recently published work in Nature Communications, we began by exploring whether microbial profiles and functional properties of duodenal biopsies from celiac patients and healthy controls were different. We observed that small intestinal biopsies from celiac patients had higher proteolytic activity towards gluten, meaning there is an increase in bacteria that can use gluten as an energy source in celiac patients, and this correlated with changes in relative abundance of microbial species with known gluten-degrading capacity (such as Pseudomonas). To determine whether the higher microbial proteolytic activity in celiac patients could induce inflammation in the host, we colonized germ-free mice with specific bacteria or communities of interest and studied the in vivo proteolytic and physiological characteristics that were induced in the colonized mice. Using a strain of P. aeruginosa with high elastase activity, and a knock-out strain that lacked elastase activity, we found that colonization of mice with the elastase-producing bacteria increased small intestinal proteolytic activity and induced innate immune activation. Notably, we saw that P. aeruginosa-derived elastase increased intraepithelial lymphocytes in the mice, a key innate immune cell type that mediates tissue damage in celiac disease. These responses were independent of gluten, and of other microbes, since similar responses were seen in mice that were mono-colonized with the elastase-producing P. aeruginosa and in mice that were maintained gluten-free. Colonization of protease-resistant PAR-2 mutant mice, or treatment with a PAR-2 antagonist, revealed that the elastase-induced immune effects were also dependent on PAR-2 signalling in the host. These experiments were all conducted in wild-type mice, that have no genetic predisposition for celiac disease. Although we observed immune activation and increases in cell types known to participate in celiac disease pathogenesis, the mice remained healthy. The next logical step was to test what would happen in a host that was genetically susceptible.
For these experiments we used mice that express the human DQ8 celiac disease susceptibly gene, and for which we have previously shown develop some characteristics of human disease when sensitized with gluten. Colonization of these genetically susceptible mice with the elastase-producing P. aeruginosa induced innate activation, similar to what was observed in the wild-type mice. However, under these conditions we determined that gluten synergized with elastase from P. aeruginosa and led to more severe inflammation and moderate small intestinal damage.
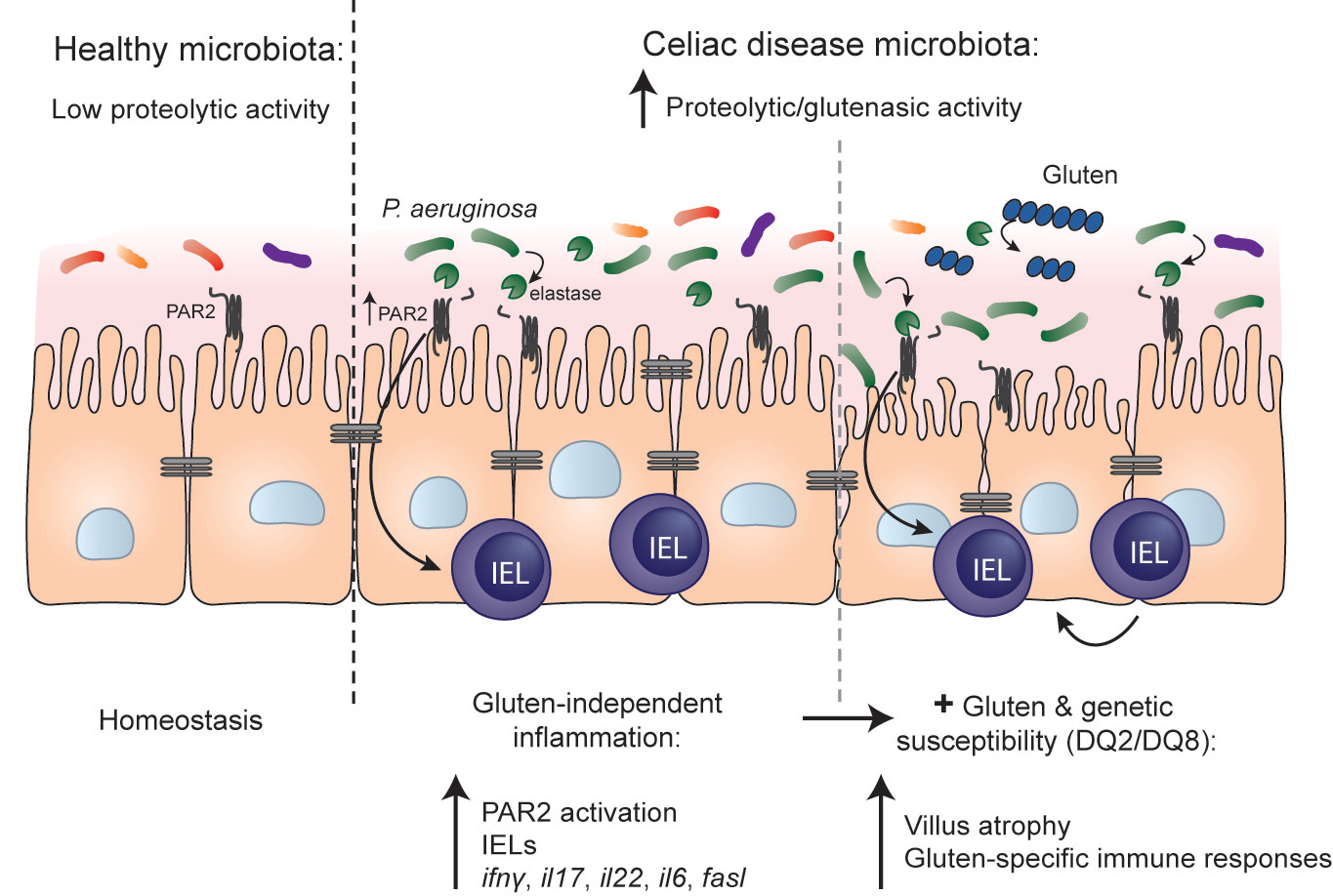
Finally, we colonized germ-free mice with aspirates from celiac patients or healthy controls to test whether whole microbial communities induced similar host responses. We found that the higher microbial proteolytic activity towards gluten in celiac patients could be transferred to mice, and this associated with higher counts of intraepithelial lymphocytes.
Microbe-modulating therapies are often used to treat inflammatory diseases without knowing the underlying mechanisms through which they exert their effect. Here we show specific mechanisms through which the activity of certain microbes can influence host responses to dietary antigens, either gluten-specific immune responses, as we previously demonstrated, or non-specific innate immune activation, both of which could increase susceptibility to food sensitivities in a genetically susceptible host. These pathways could be exploited to develop adjuvant therapies for food sensitivities, like celiac disease, or as preventative strategies in genetically susceptible individuals.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in