From a Script to GlycoGenius: How a Frustration Turned Into a Solution for Glycomics Data Analysis
Published in Protocols & Methods and Cell & Molecular Biology

Back in 2019, when I first started working with mass spectrometry analysis of glycans, I eventually hit what felt like a brick wall made of data. At that time, the standard workflow for analyzing mass spectrometry data was painfully manual. You would open the vendor software, type in the m/z values of the molecules you were interested in, trace the chromatographic peaks, check the spectra to confirm that each one was indeed monoisotopic and carried the correct charge state, and finally quantify the peak to estimate how abundant that molecule was in your sample.
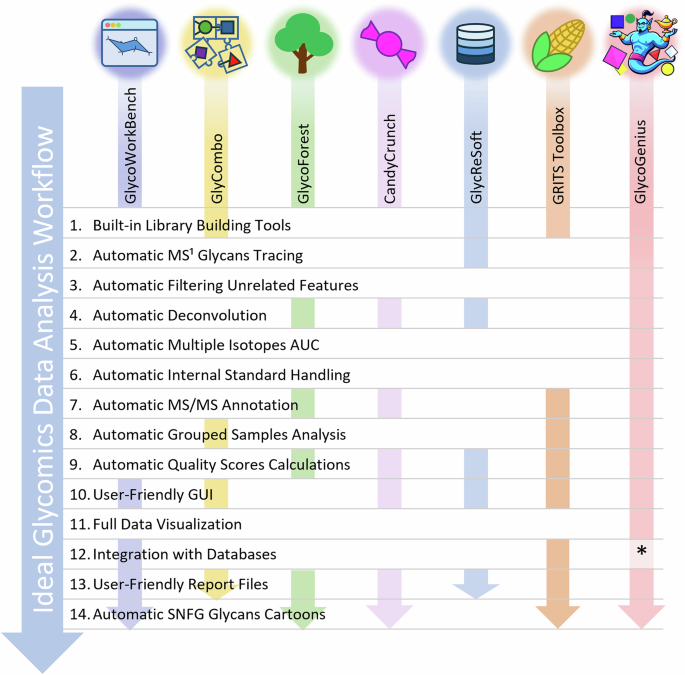
Now, imagine having to repeat that same process for hundreds of different glycans across dozens of samples. That was not just tedious; it was nearly impossible to scale. What took hours for one molecule could easily turn into months of repetitive work for a full dataset. It baffled me that such manual work was still necessary when, in other areas of mass spectrometry such as proteomics, computational tools already automated most of the workflow. Proteomics researchers had software that could take in raw data and output a neatly curated list of identifications and their abundances. Why could we not have the same for glycomics?
So, I started looking for ways to make my data analysis work easier. I tested several programs that aimed to automate glycomics data analysis, but every single one of them failed somewhere along the way. Either they could not correctly identify monoisotopic peaks, or they assigned the wrong charge states, or they produced results that needed so much manual verification that it defeated the whole purpose of automation. And even if the program got those basic steps right, there was still the deeper question: was that monoisotopic peak really the glycan we thought it was? To confirm that, we would have to look into the fragmentation spectra. It was like solving an endless puzzle, one piece of evidence at a time.
The First Spark
Out of sheer necessity, I wrote a small script to analyze my data more efficiently. It was not elegant, and it was never intended for anyone else to use, but it worked. The script processed my spectra, identified peaks, and gave me just enough information to verify my results quickly. That little script became a key part of my master’s dissertation, and during my defense, it drew quite a bit of attention. My supervisor jokingly called it GlycoGenius, a nickname that, at the time, sounded far more sophisticated than the script itself. It was not really a software; it was a collection of functions tied together by hope and caffeine.
I did not see myself as a programmer, and it was not my main goal to become one. So, after finishing my dissertation, I moved on to other projects: metabolomics, biochemical analyses, and only occasional glycomics work. GlycoGenius remained a nice memory, a clever solution for a specific problem, but nothing more.
The Idea Comes Back
Then, in 2023, an unexpected opportunity changed everything. I got the chance to spend a year abroad at the University of Groningen in the Netherlands, working with the Analytical Biochemistry group. While waiting for my samples to arrive to be analyzed there, I had some spare time. During a conversation with one of my principal investigators, a seasoned glycomics expert, I realized something surprising: the same challenges I faced in 2019 were still there. Researchers were still spending countless hours manually verifying peaks, charge states, and identifications one by one. Despite all the progress in other omics fields, glycomics was still lagging behind in terms of automated data processing.
That realization reignited the spark. Maybe that little script I wrote years ago could evolve into something much more meaningful, not just for me, but a tool for the community.
From a Script to a Tool
So, I started coding again. My goal was to build a more general-purpose, user-friendly program that could analyze multiple samples at once and automatically generate reliable identification and quantification tables. At first, it was just a command-line tool. It could process large datasets, extract key features, and summarize results in neat tables. It was functional but limited, as glycomics analysis is inherently visual. Researchers need to see their data to check peaks, confirm patterns, and compare spectra side by side to trust it.
That meant one thing: the tool needed a well-designed graphical user interface, which was a challenge I had never faced before. Fortunately, the project soon became a team effort. Colleagues joined in with ideas for the interface, feedback on usability, and suggestions based on their experience testing the program. Others shared valuable datasets that pushed the software to its limits , helping us optimize the algorithms. The visual design was discussed and polished together, combining perspectives from both experimentalists and programmers. We wanted not only something that worked but something that felt intuitive to use.
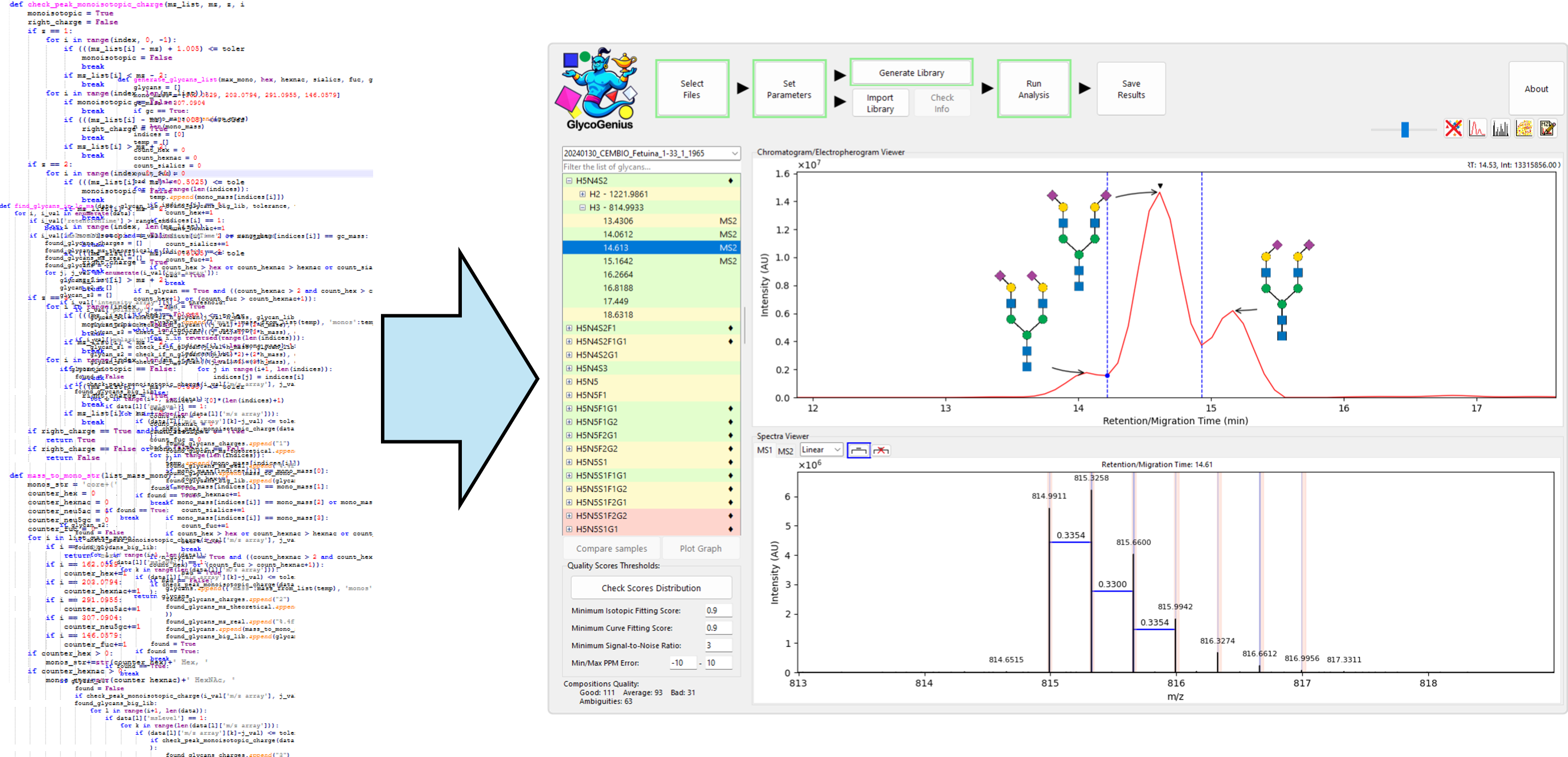
Building GlycoGenius
After many months of trial and refinement, GlycoGenius evolved into a complete analysis platform. It could guide users step by step through the workflow, visualize results, perform smart error checks, and offer flexible options for exploring annotated LC/CE-MS(/MS) data of glycans. Continuous feedback helped us polish both the performance and the design. Even the name, initially a casual nickname, became part of a unified identity for the project and defined the logo.
By then, GlycoGenius was no longer a personal project. It was a collective creation shaped by scientists, programmers, and designers who believed that glycomics deserved better analysis tool that is reliable, fast, and accessible.

Looking Ahead
Developing GlycoGenius has been an unexpected journey, growing from frustration into innovation, and from a solo script writing into a collective effort. It reminded me how small, personal problems can evolve into something that benefit an entire field when shared among people with different skills and perspectives.
There is still much to do. Fragmentation spectra and structural elucidation remain open challenges, and future versions will aim to tackle those missing aspects. As glycomics continues to expand and intersect with other omics fields, we hope GlycoGenius will help researchers spend less time on repeating routine analyses, and more time on the discoveries that truly matter.

Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in