The journey towards single-molecule protein fingerprinting
Published in Cell & Molecular Biology
In our recent manuscript published in Nature Nanotechnology (doi: 10.1038/s41565-023-01598-7)1, we developed a high-resolution fluorescence-based protein fingerprinting approach by combining DNA nanotechnology and single-molecule fluorescence. We demonstrate that we can localize multiple amino acids within a full-length protein with high precision. We further extended our technology for the detection of native proteins and a small post-translational modification, by developing new labeling approaches.
Shaping up the idea.
Ever since the start of the Chirlmin Joo lab, one of its main missions has been to contribute and develop a single-molecule protein sequencing (SMPS) platform.2,3 After the completion and publication of our first single molecule peptide fingerprinting manuscript4 I joined Chirlmin’s lab for my PhD project to work on the further development of the single molecule protein sequencer. We set out to think of a new fingerprinting approach that would eliminate the need of a protease, thereby keeping the proteins intact and would cope with the photobleaching problem to allow for long measurement times. We went back to the drawing board and the answer was; we will anchor proteins to a surface in their full-length form and obtain a structural fingerprint for each protein individually – we will pinpoint the locations of cysteines and lysines in the 3D structure of the protein using single molecule FRET (smFRET).
This fingerprinting approach brought several technical challenges of which these were considered most important to overcome: conventional smFRET experiment suffer from limited measurement times due to photobleaching and currently smFRET allows one to deal with one or two FRET pairs at a time. Therefore, we had to develop a new smFRET approach that would deal with these technical challenges.
In 2018, we were brainstorming on how to tackle these challenges. We came up with the idea of exchanging the fluorescent probes on a target protein and: FRET X, single molecule FRET via eXchange, was born!, The exchange (or switching) of fluorescent signals is a concept that was introduced, and has been key to the super-resolution community for many years already. In DNA-PAINT, this switching is governed by the rapid binding and unbinding of short DNA oligos.5,6
After several iterations of the experimental design, we finally came up with a sketch on what the eventual assay would look like that would allow for structural protein fingerprinting using FRET X (Figure 1).
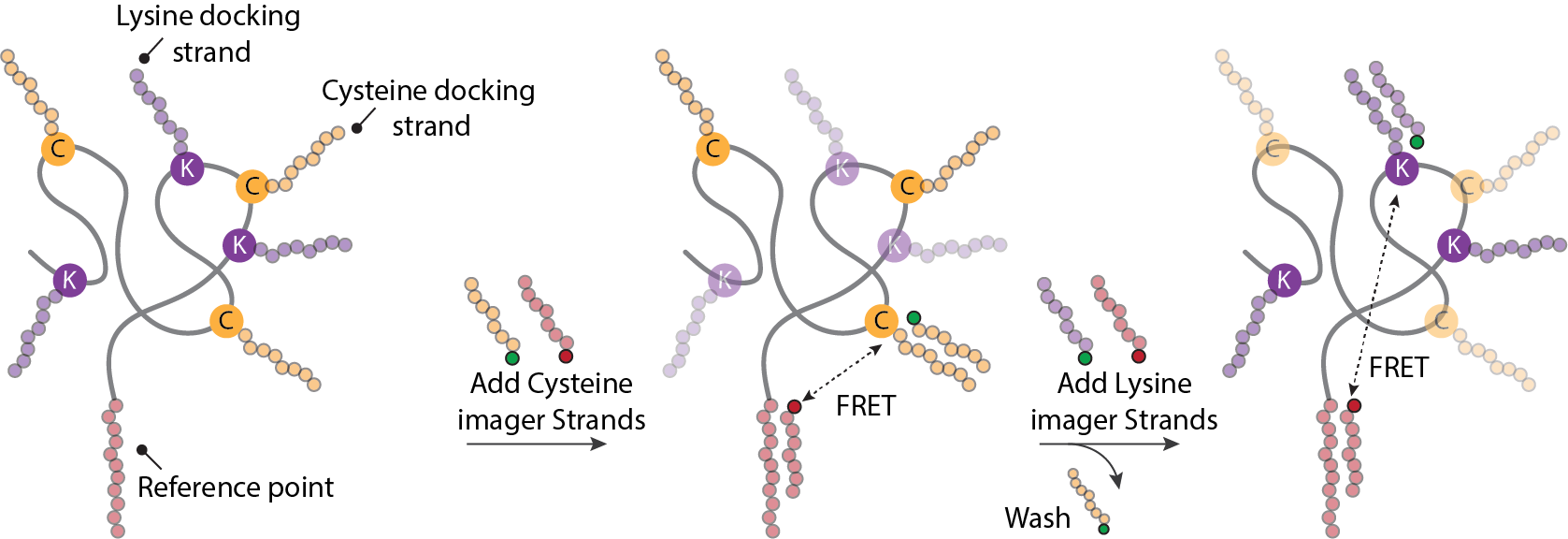
Figure 1: Schematic overview of structural protein fingerprinting using FRET X. Proteins are labelled with orthogonal DNA sequences and immobilised onto a surface. Next an imaging mixture is added with fluorescently labelled imager strands for cysteine and the reference point. After obtaining sufficient FRET binding events for each of the cysteine, the chip is washed and imaging mixture for lysines is added. This cycle can be repeated for any labelled amino acid or post translational modification within a protein.
The idea was relatively straightforward, we would label different amino acids (cysteines and lysines) with orthogonal DNA sequences and determine their relative position to a single, unique reference point – that is another orthogonal DNA strand attached to either the N or C terminus of the protein. The reference point would also function as the protein anchor to allow for the proteins to be attached to our surface. Then by injecting imager strands for the reference point + imager strands for the cysteines, we would determine the relative position of each cysteine in the protein structure first (being part of the fingerprint), then wash the system and repeat this for lysines or any other amino acid or PTM of interest. This design allowed us to control the exchange of DNA-labelled fluorescent probes (or imager strands) by altering the length and sequence of the DNA oligos, this would mean that under optimal experimental conditions we would only measure a single FRET pair at a time (even when many are present) and the binding/unbinding cycle of the imager strands can go on forever – thereby eliminating photobleaching issues. Thus on paper we had overcome the two biggest challenges.
Paving the way for FRET X based protein fingerprinting.
As any experimentalist will know, ideas that work on paper and experiments that work in the lab can be worlds apart. To evaluate the potential of FRET X for protein fingerprinting, we set out to answer the following questions: 1) Does our FRET X technology really allow us to measure multiple points of interest in a single molecule and can we indeed eliminate the photobleaching issue? and 2) Is the structural information generated sufficient for a fingerprint to be unique for >90% of the proteome?
We set out to answer the second question using DNA constructs only. Why DNA?, you might ask, well that is because it allowed us to have full control over the experimental design (we could pretty much order every crazy construct that we wanted) and answer all sorts of fundamental questions of the FRET X technology. The experiments on DNA constructs showed encouraging results, which were published in 2021.7
During that same time, we reached out to Dick de Ridder in Wageningen University where Carlos de Lannoy helped us answer the second question using bioinformatic approaches. Carlos had obtained very promising results on how unique a structural fingerprint would be for the majority of the proteome. They had further extended their work in developing a lattice model that could predict what the FRET X fingerprint of proteins would look like. Having demonstrated the method on DNA, we took it to the next level: short synthetic peptides. This came with some hurdles in labelling and purification, but eventually the experimental data nicely agreed with the predicted fingerprints.8
Now it was time to move on to the big leagues: working with proteins! Over the years I have developed a love/hate relationship with these molecules. They are fascinating molecules in their shapes and functions, but can be a nightmare to work with! Therefore, we started working with a simple protein (that has no structure) alpha synuclein to further challenge the FRET X technology. After obtaining the first promising results, we increased the complexity and selected several biomarkers that were successfully fingerprinted using FRET X. Finally, we further expanded the FRET X technology to allow for the detection and localization of post translational modification in individual, full-length proteins.
Final Thoughts
Altogether, there were many lessons learned during this project, both scientific lessons and personal. First and most important; I’d like to think as science as a team sport, this (and many other) projects would not have been possible without the incredible multidisciplinary team that I had the pleasure and honour of working with. Secondly, I think one of the key things to make this project a success is that we started and built this project from the ground up, meaning that we started very simple and slowly increased the complexity to peptides and ultimately to the detection of proteins. Because performing scientific experiments is simple – but designing simple, meaningful experiments is one of the most difficult things to do!
References
- Filius, M. et al. Full-length single-molecule protein fingerprinting. Nat. Nanotechnol. 1–8 (2024) doi:10.1038/s41565-023-01598-7.
- Restrepo-Pérez, L., Joo, C. & Dekker, C. Paving the way to single-molecule protein sequencing. Nature Nanotechnology 13, 786–796 (2018).
- Alfaro, J. A. et al. The emerging landscape of single-molecule protein sequencing technologies. Nature Methods 18, 604–617 (2021).
- Ginkel, J. V. et al. Single-molecule peptide fingerprinting. 1–6 (2018) doi:10.1073/pnas.1707207115.
- Schnitzbauer, J., Strauss, M. T., Schlichthaerle, T., Schueder, F. & Jungmann, R. Super-resolution microscopy with DNA-PAINT. Nature Protocols 12, 1198–1228 (2017).
- Van Wee, R., Filius, M. & Joo, C. Completing the canvas: advances and challenges for DNA-PAINT super-resolution imaging. Trends in Biochemical Sciences (2021) doi:10.1016/j.tibs.2021.05.010.
- Filius, M., Kim, S. H., Severins, I. & Joo, C. High-Resolution Single-Molecule FRET via DNA eXchange (FRET X). Nano Lett. 21, 3295–3301 (2021).
- Lannoy, C. V. de, Filius, M., Wee, R. van, Joo, C. & Ridder, D. de. Evaluation of FRET X for single-molecule protein fingerprinting. iScience 24, 103239 (2021).
Follow the Topic
-
Nature Nanotechnology

An interdisciplinary journal that publishes papers of the highest quality and significance in all areas of nanoscience and nanotechnology.




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in