HIV affects polyamine metabolism and T cell lineages
Published in Healthcare & Nursing
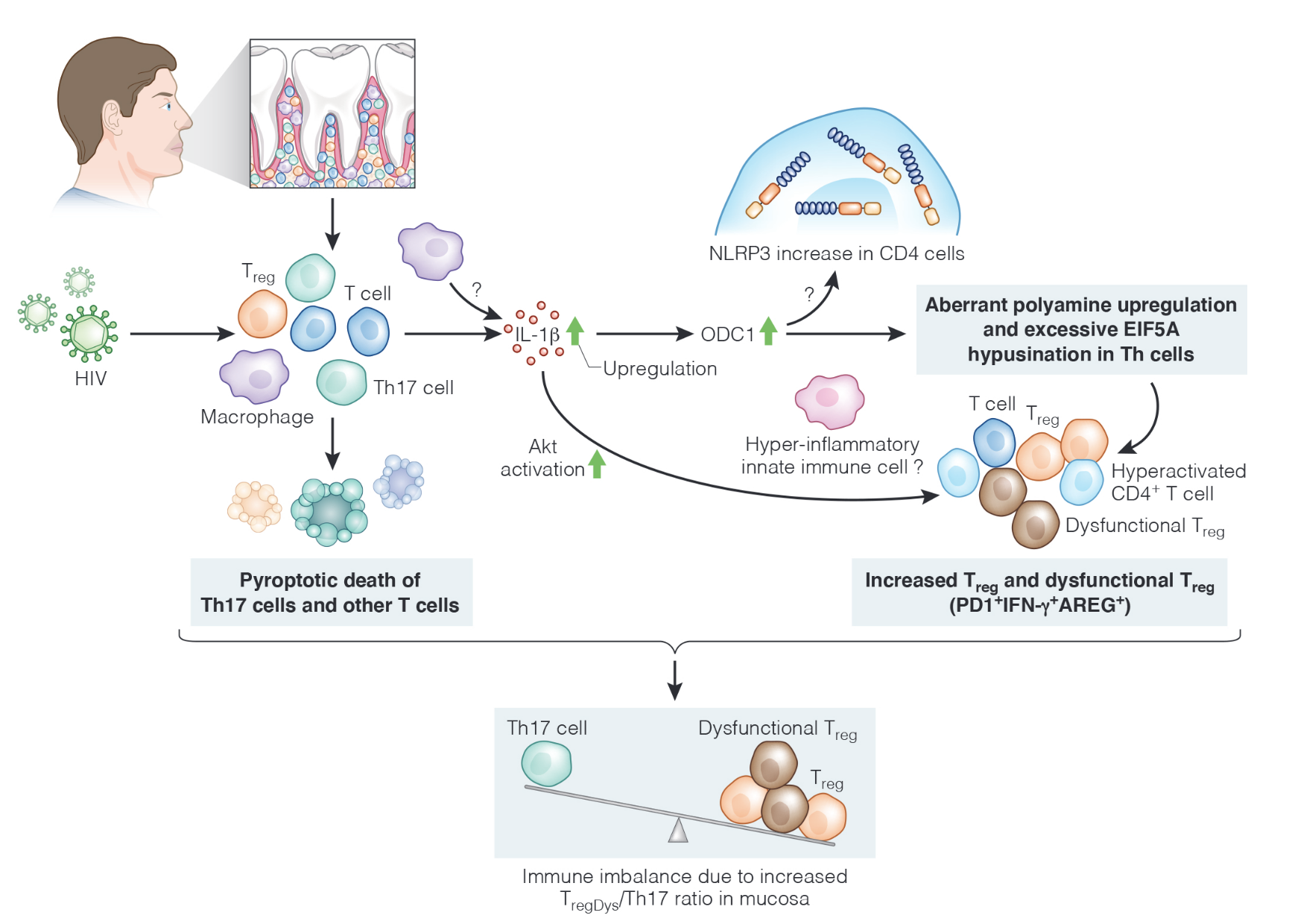
Polyamines are organic polycations that work together with DNA, RNA, and proteins, and control cell growth and death. Polyamines come from food and can be produced by our cells and the microbiome in our mucosa. Aberrant changes in their levels in cells are associated with aging and diseases. A study by Dr. Pandiyan's team at Case Western Reserve University has explored polyamines' role in HIV infection and has shown that HIV could damage our immune system by increasing T-cell polyamine metabolism. (Nature Communications; DOI 10.1038/s41467-023-36163-2; https://doi.org/10.1038/s41467-023-36163-2 or https://www.nature.com/articles/s41467-023-36163-2.pdf).
Mechanistic studies using an in vitro human tonsil organoid infection model revealed that HIV infection of T cells also resulted in increased polyamine synthesis, which was dependent on the activities of caspase-1, IL-1β, and ornithine decarboxylase-1 (ODC-1). HIV-1 also led to a heightened expression of polyamine synthesis intermediates including ODC-1 as well as an elevated dysfunctional regulatory T cell (TregDys) /T helper 17 (Th17) cell ratios. The loss of Th17 cells was due to pyroptotic cell death. Blockade of caspase-1 and polyamine synthesis intermediates reversed the TregDys phenotype showing the direct role of polyamine pathway in altering T cell functions during HIV-1 infection. Lastly, oral mucosal TregDys/Th17 ratios and CD4 hyperactivation positively correlated with salivary putrescine levels, which were found to be elevated in the saliva of HIV+ patients. The anti- or hyper-inflammatory effect of altered polyamines on other innate immune cells and the role of increased NLRP3 expression remain to be studied in the future. Thus, by revealing the role of excessive polyamine synthesis during HIV infection, this study unveils a mechanism by which chronic viral infections could drive distinct T cell effector programs and Treg dysfunction.
This study could lead to new therapeutic approaches to repair the immune system that is damaged due to aging, chronic infections, inflammation, and cancer.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in