How to build a flagellum for all conditions
Published in Microbiology
When starting his PhD some years ago, Marco Kühn observed that our model species, monopolarly-flagellated Shewanella putrefaciens, got stuck in narrow passages but escaped the trap by wrapping their flagellar filaments around their cell bodies and backing out in a screw-like fashion (see here). In a great collaboration with Felix Schmidt and Bruno Eckhardt from the Physics Department at Marburg University we jointly showed that flagellar screw formation is triggered at high load due to an instability of the flagellar filament close to the cell body. Thus, these bacteria exploit the mechanistic properties of the flagellar filament for a novel type of movement, which was further explored in this study, for which three groups joined forces. Combining the expertise of these groups, we could comprehensively study the swimming abilities of Shewanella and link it to the composition of the flagellum. The story can be found here.
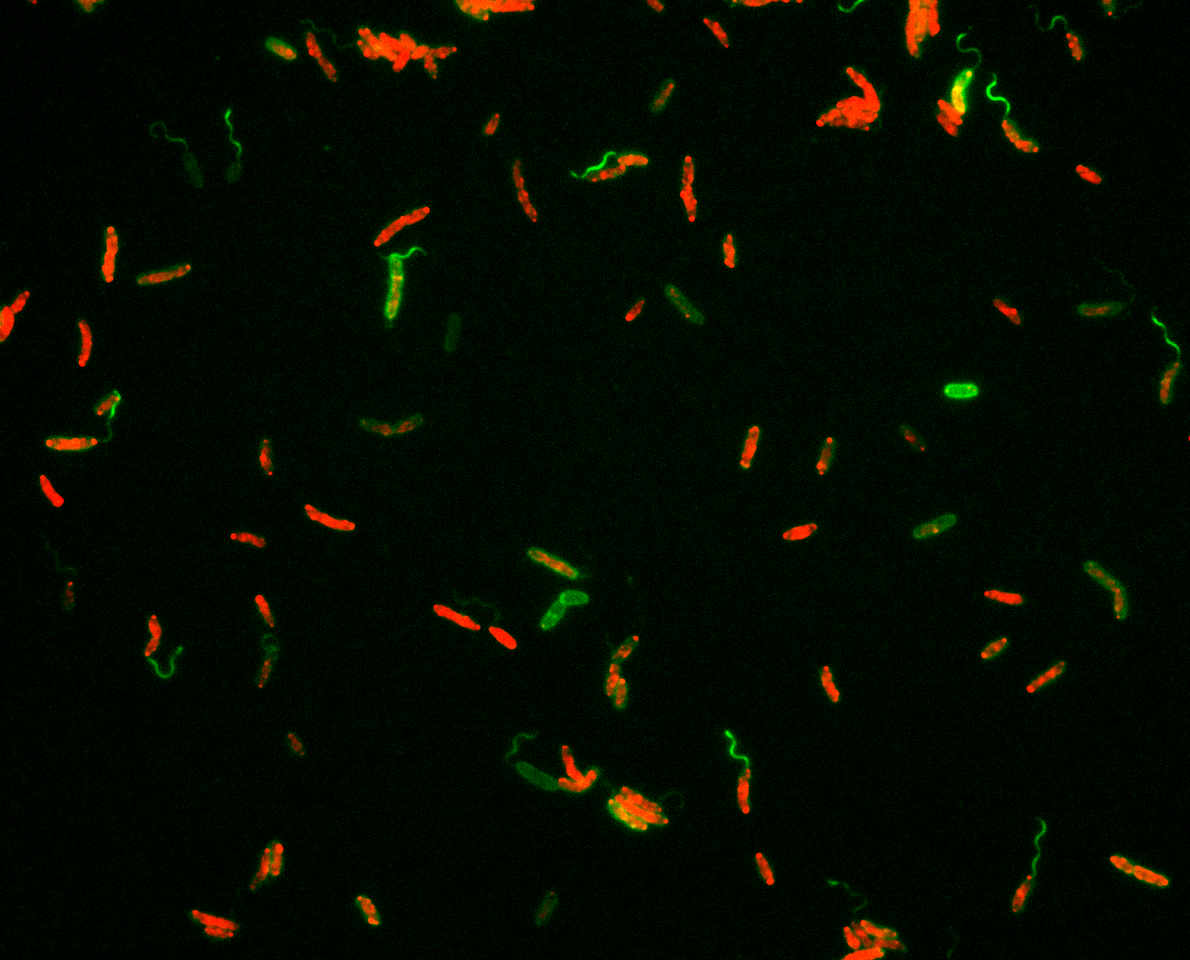
In about half of all flagellated bacterial species, the flagellar filament is composed of more than one individual building block, the flagellin. So far there is little evidence for why so many bacteria maintain this setup: more components means more things that could go wrong, and more efforts required to maintain them. In the case of Shewanella, the flagellum is formed by two flagellins, FlaA and FlaB. By fluorescently labeling the individual flagellins, we found that they are spatially organized within the filament with FlaA mainly occurring in a proximal segment close to the cell body and FlaB making up the remaining length (Fig. 1). By mutant studies, we explored the underlying regulatory mechanism (the two flagellins are sequentially exported) and produced strains in which the flagellar filament solely consisted of FlaA or FlaB, or where FlaB is located at the base of the filament.
But how do cells move with these different flagellar filaments? To answer this, Nicola Farthing and Laurence Wilson in York used their fantastic set-up to trace swimming cells in 3D. They showed that there are indeed significant differences in some parameters, such as swimming speed or turning angles. However, all cells, wild type as well as flagellar mutants, were generally able to swim well in ‘open waters’. The same strains were also tested for their ability to move through soft agar, representing a highly structured environment. Somewhat unexpectedly, the cells’ ability to spread through soft agar did not correlate well with the ability of free swimming: FlaA-only filamented cells completely outperformed FlaB-only mutants in free swimming, but were severely restricted in spreading, whereas the FlaB mutant performed almost as well as wild-type cells in soft agar (Fig. 1).
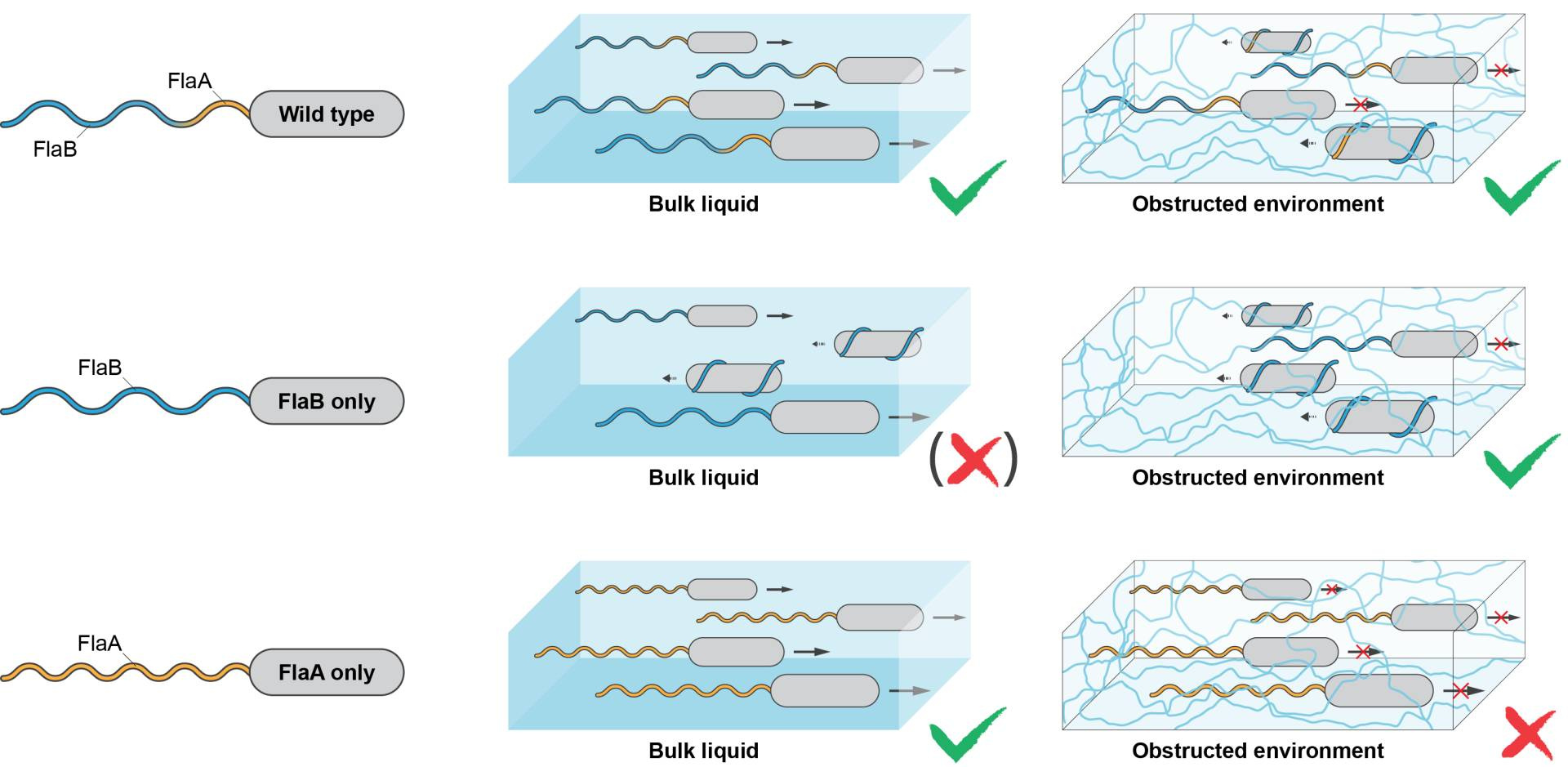
Figure 1: Building a flagellar filament for all conditions. The wild type (upper panel) forms a filament by FlaA and FlaB in spatial arrangement. This stabilizes the filament sufficiently to allow swimming but also to form flagellar screws in structured environments, which increases spreading under these conditions. FlaB-only filamented cells (middle panel) have destabilized flagella, which is disadvantageous for free swimming (but works well in structured environments). FlaA-only filamented cells (lower panel) have rather stable filaments, which results in good free swimming but causes a massive decrease in spreading through structured environments.
Microscopic studies on these strains showed that this difference in spreading was mainly due to the inability of FlaA mutants to wrap the flagellar filament around the cell and to perform the screwing mode of movement. By performing numerical simulations, Felix Schmidt and Bruno Eckhardt provided a neat explanation: the proximal FlaA segment stabilizes the base of the filament against premature screw formation, which was in agreement with all experimental observations. Notably, the wild-type flagellin set-up provides a very good trade-off between stability for free swimming while maintaining the ability to form a screw.
Thus, in this joint study we could show that bacteria may maintain the multi-flagellin set-up to enable fine tuning of the flagellar filament’s mechanistic properties (a nice example for functional redundancy in biological systems) to adjust swimming and spreading capacities. In addition, screw formation has now been observed for a number of polarly flagellated bacteria other than Shewanella, indicating that the screwing mode of movement is widespread among bacterial species. Our study now provides solid evidence that screwing motility is in fact beneficial in numerous biological settings where bacteria have to move through structured environments.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in
The 'Read the paper' button links to the old PNAS paper (2017).
The Nature Communications article can be found here:
Spatial arrangement of several flagellins within bacterial flagella improves motility in different environments
https://www.nature.com/articles/s41467-018-07802-w