Hydrogen production by Epsilonproteobacteria involved in methane-producing co-cultures
Published in Microbiology

The original article was published on Nov 19 2018 in Nature Communications
The beginnings: hydrogen production in Epsilonproteobacteria?
Our investigations on hydrogen production in Sulfurospirillum spp., free-living Epsilonproteobacteria (note that Epsilonproteobacteria were recently suggested to be reclassified and renamed to Campylobacterota) had its roots in the genomics of the organohalide-respiring Sulfurospirillum multivorans. I
wondered why the genome contained four hydrogenase gene clusters,
since most Epsilonproteobacteria (like the well-known pathogenic
Campylobacter jejuni and Helicobacter pylori) were known for they simple hydrogen metabolism: a one-way for
hydrogen uptake, dependent on only one periplasmic, membrane-bound
hydrogenase. Also the non-pathogenic Sulfurospirillum spp., mostly known for anaerobic respiration with ‘exotic’, often toxic electron
acceptors like arsenate, selenate and, in the case of S. multivorans, organohalides such as tetrachloroethene were described to use hydrogen as electron donor. Hydrogen consumption was very similar to that of C. jejuni and H. pylori via a hydrogen-uptake hydrogenase, as characterized earlier by us. However, one of the four hydrogenase
gene clusters seemed to code for a membrane-bound hydrogen-evolving
hydrogenase, and since the organism was described to ferment pyruvate
when no electron acceptor was provided, which often facilitates
hydrogen production in other bacteria, I convinced our doctoral
student Stefan Kruse (Figure 1) to do some experiments on pyruvate fermentation
in several different Sulfurospirillum spp. Our goal from the
beginning on was to establish Sulfurospirillum spp. in syntrophic
relationships as a hydrogen producer.

Two surprising findings: 1) adaptation to fermentation and 2) different hydrogen producing capacities in Sulfurospirillum cavolei and Sulfurospirillum multivorans
The first
surprise was that the bacteria adapated to pyruvate fermentation –
at first growth was quite slow, but after several transfers on
pyruvate without electron acceptor, the bacteria seemed to accept
their fate and grew nearly as good as under respiratory metabolism.
While these results were scientific “by-products” back then and only included
briefly in the biorxiv.org preprint published nearly one year ago, a
reviewer convinced us to do proteomics (in collaboration with Lorenz Adrian of the Helmholtz Center for Environmental Research) of adapted and non-adapted
cells during the revision process after submitting to Nature Communications. Thus, we showed that
this adaptation depended on only a few proteins with a role on which only can be speculated. The second surprise arose when we compared
different Sulfurospirillum spp. and observed a much higher
hydrogen production in Sulfurospirillum cavolei. A genomic
comparison between the two Sulfurospirillum spp. led us on
a wrong track initially, which Stefan Kruse presented in a talk at
the renowned hydrogenase conference, 2015 held in Marseille (France):
S. cavolei encodes a hydrogenase with an iron-only active
center and these hydrogenases are known for hydrogen production in
many bacteria – so we thought this iron-only hydrogenase is responsible for the difference in hydrogen production. However, we were puzzled by two observations, 1) there was a mutation in the S. cavolei iron hydrogenase gene cluster and 2) S. multivorans and S. cavolei produced different organic acids during fermentation. Again, proteomics and genomics, combined with a careful analysis of the HPLC results of fermentation products, aided us. Instead of the iron hydrogenase protein, we found only one large (eight subunits) membrane-bound, cytoplasmic nickel-containing hydrogenase in S. cavolei, the same as in S. multivorans, so differences in hydrogenase equipment was not the reason for the difference in hydrogen production. Instead it was rooted in different different fermentation metabolisms causing S. multivorans producing succinate and lactate additionally to acetate, consuming electrons which are channelled into hydrogen production in S. cavolei.
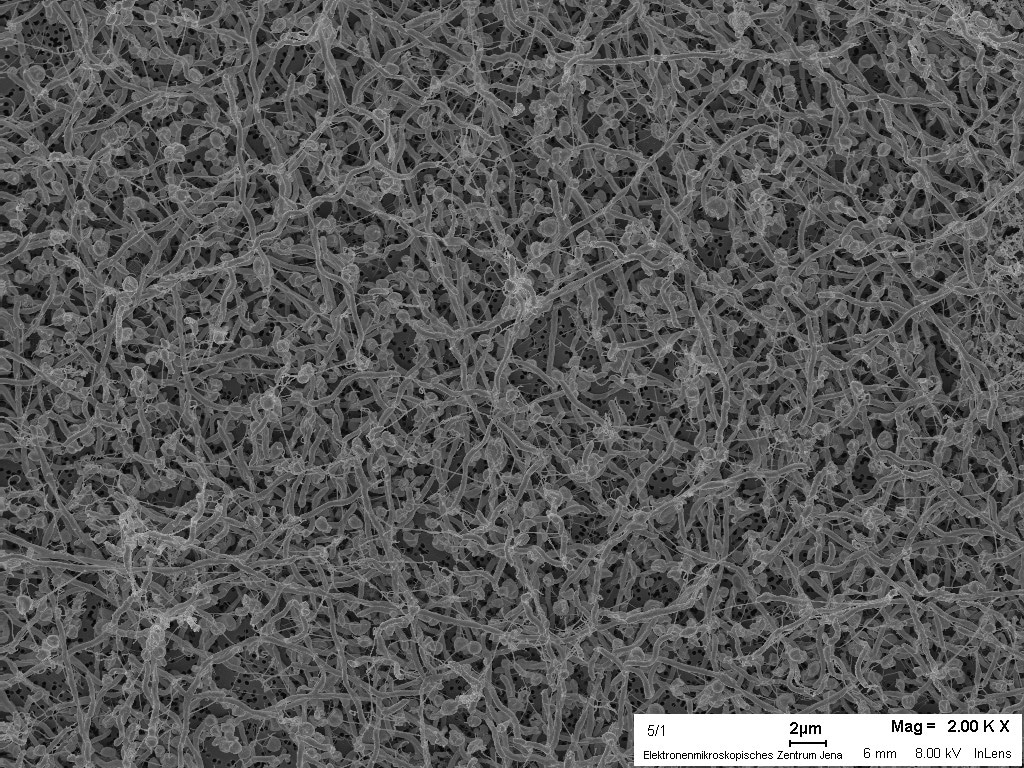
Production of methane from lactate: a syntrophic relationship of Sulfurospirillum multivorans with a methanogen
What would be the ecological relevance of this novel physiological
trait in Epsilonproteobacteria? Hydrogen-producing bacteria often
deliver hydrogen derived from fermentation to hydrogen consumers in
anoxic environments. Sometimes, the former can only derive energy
from fermentation when the evolved hydrogen is immediately consumed
by the other bacteria. This bacterial symbiosis is called syntrophy
and we found out that indeed Sulfurospirillum spp. can live
syntrophically with the hydrogen-consuming methanogenic archaeon
Methanococcus voltae, when lactate was present as single
energy substrate (Figure 2 and 3). Together with the finding of the
hydrogen-evolving hydrogenase in other epsilonproteobacterial genomes, this hints to a new ecological role of
these bacterial subclass.

The next frontiers: hydrogenase as a proton pump and the ecological and environmental impact of epsilonproteobacterial hydrogen production
Since some of the membrane-integral subunits of the hydrogen-producing hydrogenase of Sulfurospirillum spp. are related to those of the respiratory complex I (shown extensively from page 14 in the supplement of our Nature Communications publication), also bearing conserved amino acids critical for proton pumping, we assume that this hydrogenase might contribute to energy conservation, pumping protons while channeling electrons from reduced ferredoxin to protons. During the revision of our manuscript, a ground-breaking paper on the structure on the related MBH of Pyrococcus furiosus was published, which is a great help in the further investigation on such hydrogenases.
The impact of hydrogen production by Sulfurospirillum spp. on an ecological and/or environmental and biotechnological scale is also subject to further investigations. Sulfurospirillum spp. are typically found, often in high amounts, in areas rich in sulfur compounds, bioelectrodes, and the toxic electron acceptors arsenate, selenate, chlorinated ethenes. When lactate or pyruvate is present in these environments, sub-populations of Sulfurospirillum spp. might produce hydrogen when electron acceptor concentration is low in the corresponding micro-environment. Of great interest for bioremediation is hydrogen-production of the tetrachloroethene-respiring Sulfurospirillum multivorans, since other organohalide-respiring bacteria are obligate hydrogen-consumers, making an efficient and fast detoxification of tetrachloroethene to ethene feasible.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in