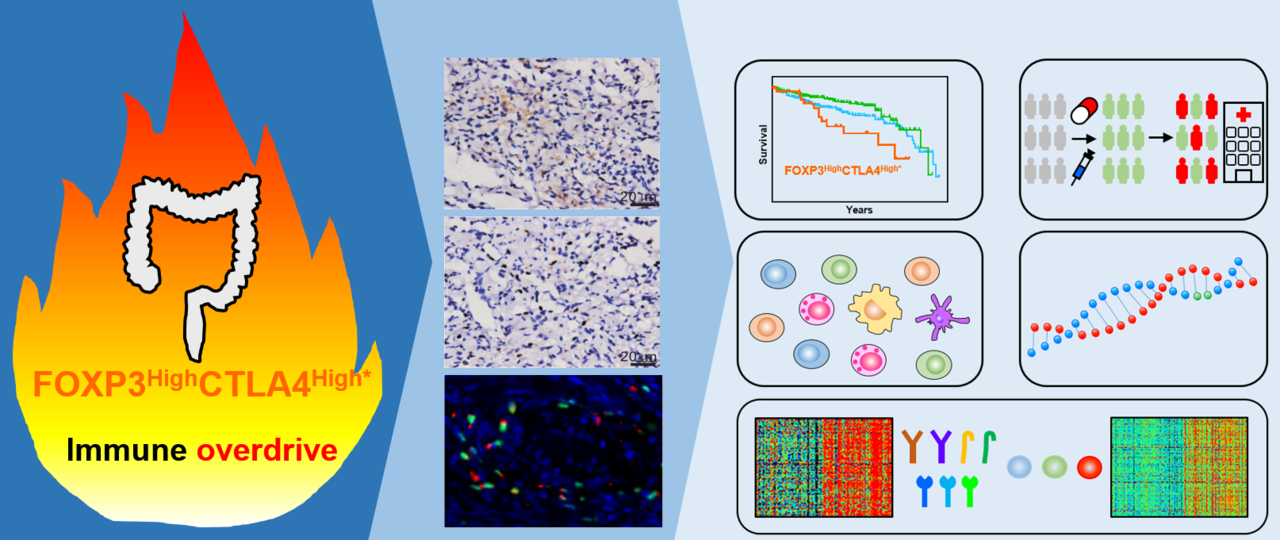
Colorectal cancer (CRC) is a solid cancer type with a complex and heterogeneous tumor microenvironment (TME). Interestingly, CRC showed some "abnormal" phenomena. For example, prior studies have revealed that a high level of immune cell signatures predicted good survival in many cancer types except CRC. Furthermore, immune checkpoints (ICs) genes often shows tumor-promoting functions in many cancer types, but predicted better survival in CRC. Thus, the identification of novel CRC molecular subtypes with appropriate TME features and prognoses is urgently desired for its precise treatment.
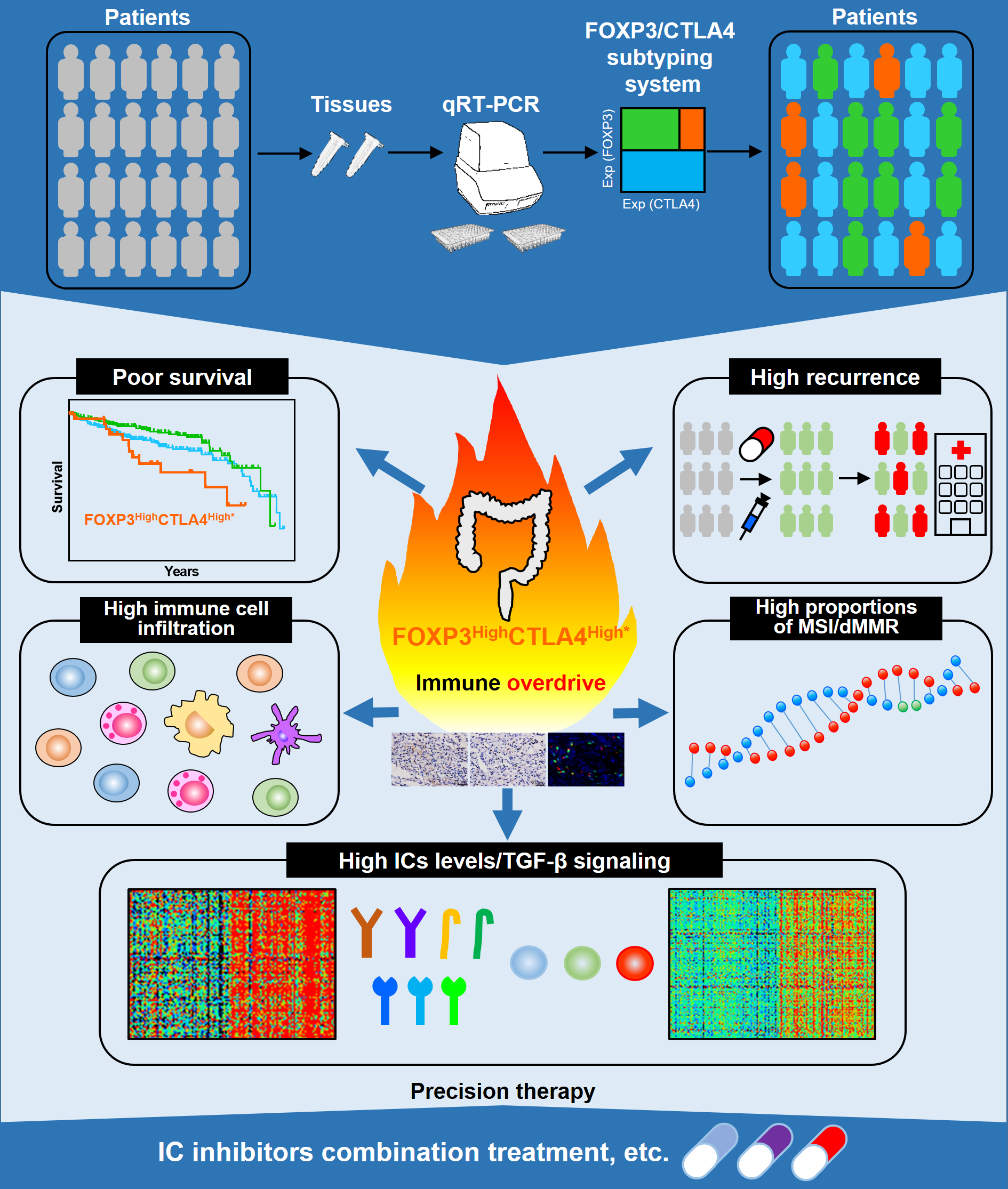
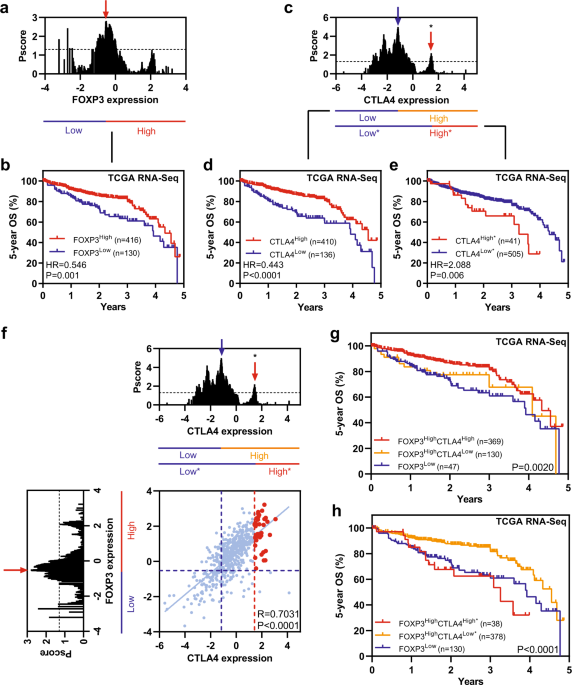
CTLA4 is mainly expressed on regulatory T cells (Tregs), which are characterized by FOXP3 expression [1]. This led our team to explore the exact roles of CTLA4 and FOXP3 in CRC. Dr. Kaisa Cui joined our team in November 2019, he is very interested in cancer bioinformatics. We used his method publishing in a previous study to analyze FOXP3 and CTLA4 survival in CRC [2]. Consistent with prior studies, both FOXP3High and CTLA4High patients showed favorable overall survival in CRC according to the best cut-off. Intriguingly, a recent study in CRC identified a highest-risk CRC subtype showing high CD8A coupled with intense CD274 [3]. This finding induced us to analyze the effects of each cut-off of FOXP3 and CTLA4 expression on survival of CRC. Interestingly, we found there are two different subpopulations with opposite survival patterns according to CLTA4 expression in FOXP3High CRC patients. Furthermore, this method was established applied to more cohorts and we identified a unique high-risk FOXP3HighCTLA4High* subpopulation in CRC. This unique risk subtype shows an immune overdrive TME phenotype, as exemplified by high levels of immune infiltration, high ICs levels, high proportions of MSI/dMMR and activation of TGF-β signaling. Additional, FOXP3HighCTLA4High* high-risk subpopulation was observed not only in CRC but also in other cancer types, including kidney renal clear cell carcinoma, kidney renal papillary cell carcinoma and low-grade glioma.
Impressively, our finding is similar to the immune overdrive phenotype described in a previous study [3]. Besides, after submitting this manuscript to Oncogene, we found a newly published paper that identified a CD8AhighIDO1high high-risk subgroup featured by high levels of the immune response, IC genes, and Th1/IFN-g gene signatures in colon cancer [4]. This study further enhanced our confidence in our research.
Immunoscore studies demonstrate that a high level of immune cell infiltration was significantly associated with improved survival in CRC [5, 6]. However, based on our study and the abovementioned researches, immune overdrive subgroups with high immune infiltration features in CRC were associated with poor survival outcomes. These exciting studies suggest that relying on a single immune evaluation may lead to the misclassification of risk in CRC.
In conclusion, the FOXP3HighCTLA4High* shows poor clinical outcomes and unique TME features with high immune cell infiltration, high levels of ICs and high proportions of MSI/dMMR, suggesting that anti-tumor immune responses are pre-existed in TME of these tumor tissues. Hence, patients of these immune overdrive subpopulations in CRC and certain cancer types may benefit from more active monitoring and treatment, especially with IC inhibitors combination treatment and in favor of developing related therapeutic strategies.
References
1 Riera-Domingo C, Audige A, Granja S, Cheng WC, Ho PC, Baltazar F et al. Immunity, Hypoxia, and Metabolism-the Menage a Trois of Cancer: Implications for Immunotherapy. Physiol Rev. 2020; 100: 1-102.
2 Cui K, Liu C, Li X, Zhang Q, Li Y. Comprehensive characterization of the rRNA metabolism-related genes in human cancer. Oncogene. 2020; 39: 786-800.
3 Fakih M, Ouyang C, Wang C, Tu TY, Gozo MC, Cho M et al. Immune overdrive signature in colorectal tumor subset predicts poor clinical outcome. J Clin Invest. 2019; 129: 4464-4476.
4 Zhang R, Li T, Wang W, Gan W, Lv S, Zeng Z et al. Indoleamine 2, 3-Dioxygenase 1 and CD8 Expression Profiling Revealed an Immunological Subtype of Colon Cancer With a Poor Prognosis. Front Oncol. 2020; 10: 594098.
5 Mlecnik B, Bindea G, Angell HK, Maby P, Angelova M, Tougeron D et al. Integrative Analyses of Colorectal Cancer Show Immunoscore Is a Stronger Predictor of Patient Survival Than Microsatellite Instability. Immunity. 2016; 44: 698-711.
6 Pages F, Mlecnik B, Marliot F, Bindea G, Ou FS, Bifulco C et al. International validation of the consensus Immunoscore for the classification of colon cancer: a prognostic and accuracy study. Lancet. 2018; 391: 2128-2139.
Follow the Topic
-
Oncogene

This journal aims to make substantial advances in our knowledge of processes that contribute to cancer by publishing outstanding research.



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in