Innophore's 3D point clouds provide a head start in monitoring emerging SARS-CoV-2 variants
Published in Healthcare & Nursing

If the COVID-19 pandemic taught us two things, it might be that flexibility is key in all ways of life and that rapidly evolving viruses call for fast evaluation systems allowing us to identify variants of concern quickly and monitor their development to buy us time to react.
With structural biology and bioinformatics aiming to keep up with the speed of evolving viruses, we took on the challenge and applied the earlier-mentioned flexibility to a previously developed technology: the use of point clouds for the characterization of binding sites.
Originally coming from the field of structural bioinformatics, we published our Catalophore™ technology[1] in Nature Communications some years ago. Since then, we have continued to develop, extend and tailor this 3D-descriptor approach to various biotech and healthtech applications. For example, at the onset of the COVID-19 pandemic, in January 2020, we contributed to the very early characterization of SARS-CoV-2 proteins mentioned in the articles “Open for outbreaks” and “The pandemic pipeline” in Nature Biotechnology.

© Innophore, January 2020
Our very first structural models have been downloaded tens of thousands of times worldwide and were used for instance by the Chinese Center for Disease Control and Prevention or by AI startups like Insilico Medicine. Shortly thereafter we joined a partnership with Harvard Medical School that provided the first large-scale drug screening[2] against 17 target proteins of SARS-CoV-2 published in 2021.
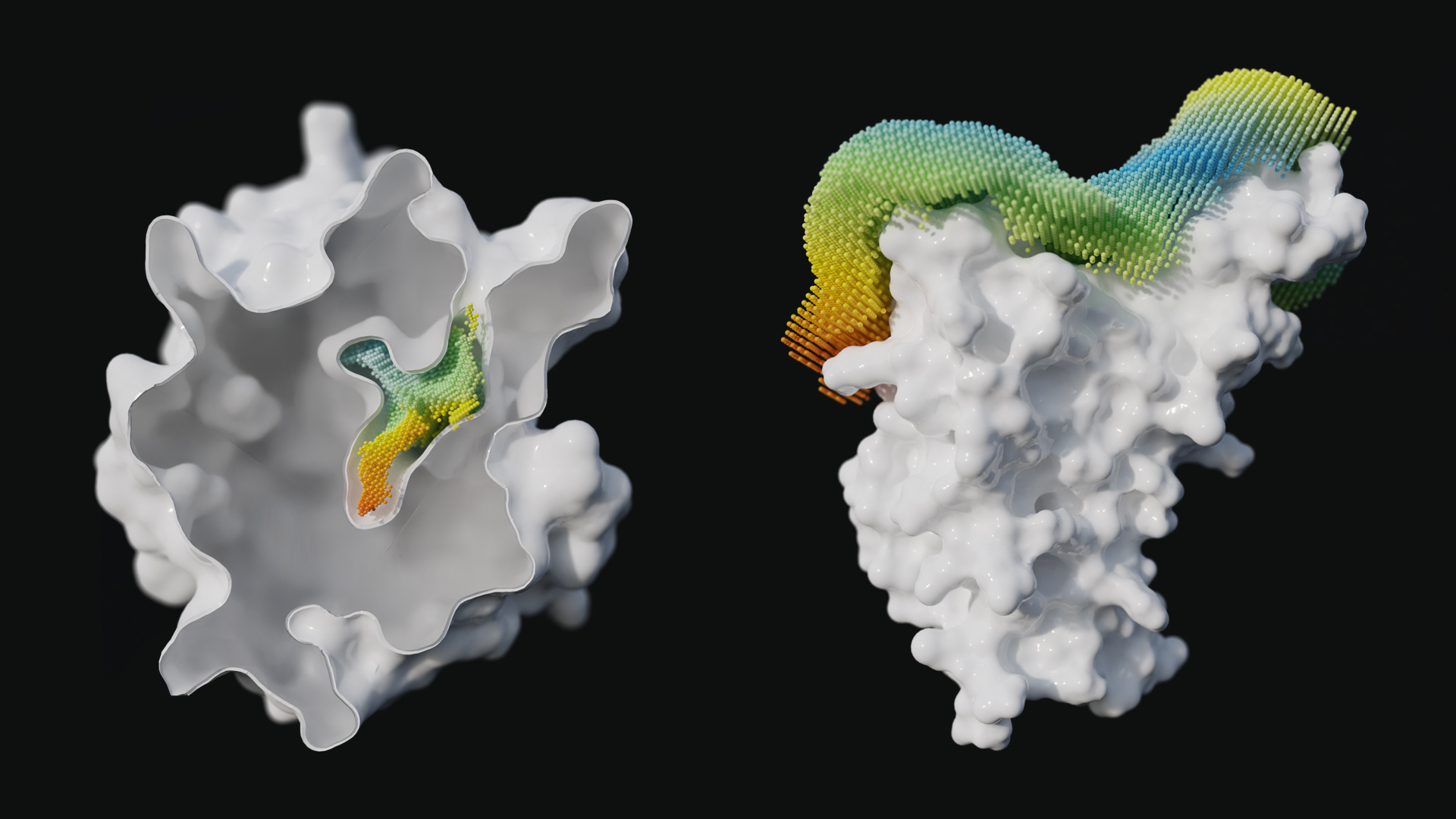
We initially used our point clouds (so called “catalophores”) to characterize binding cavities in proteins, but during the start of the pandemic, we quickly realized that they can be expanded to other areas in proteins as well: surfaces. And this is a short summary of how Halos were born.
As the pandemic evolved, it became clear that it was critical to rapidly monitor the genetic drift of the virus. Our first results were published one year ago[3] in the special issue COVID -19 of Scientific Reports. Therein we predicted a single SARS-CoV-2 spike RBD mutation S477N to increase affinity before it became dominant in New York City. Recently, Uğur Şahin, CEO of BioNTech, which has developed one of the most important vaccines against COVID -19, referred to our article in his call for further large-scale analysis of SARS-CoV-2 spike glycoprotein mutants in PLOS ONE[4].
To enable massive global structural screening of all emerging variants, we have teamed up with Amazon Web Services, the world's largest cloud provider, which has committed enormous computing resources. Now, let us guide you through the process on how we aim to predict variants of concern faster in the future.
Phase 1: Observe
Following the timeline of the waves and the distribution of variants we can see several variants emerging and significantly dominating the global pandemic event, namely the variants of concern Alpha, Beta, Gamma and Delta affecting millions of people[5] [6]. With the rise of Omicron, we decided to monitor the pandemic on a molecular level and performed a sequence and structural-bioinformatics analysis to estimate the effects of amino acid substitutions on the affinity of the SARS-CoV-2 spike receptor binding domain (RBD) to the human receptor hACE2. Previous studies indicate that increased spike-hACE2 affinity correlates with higher infectiousness[7] [8] [9].
Omicron features a high number of mutations throughout the viral genome, 39 of which cause changes in the amino acid sequence of the spike protein[10] [11] [12] making it a suspicious candidate from the get go.
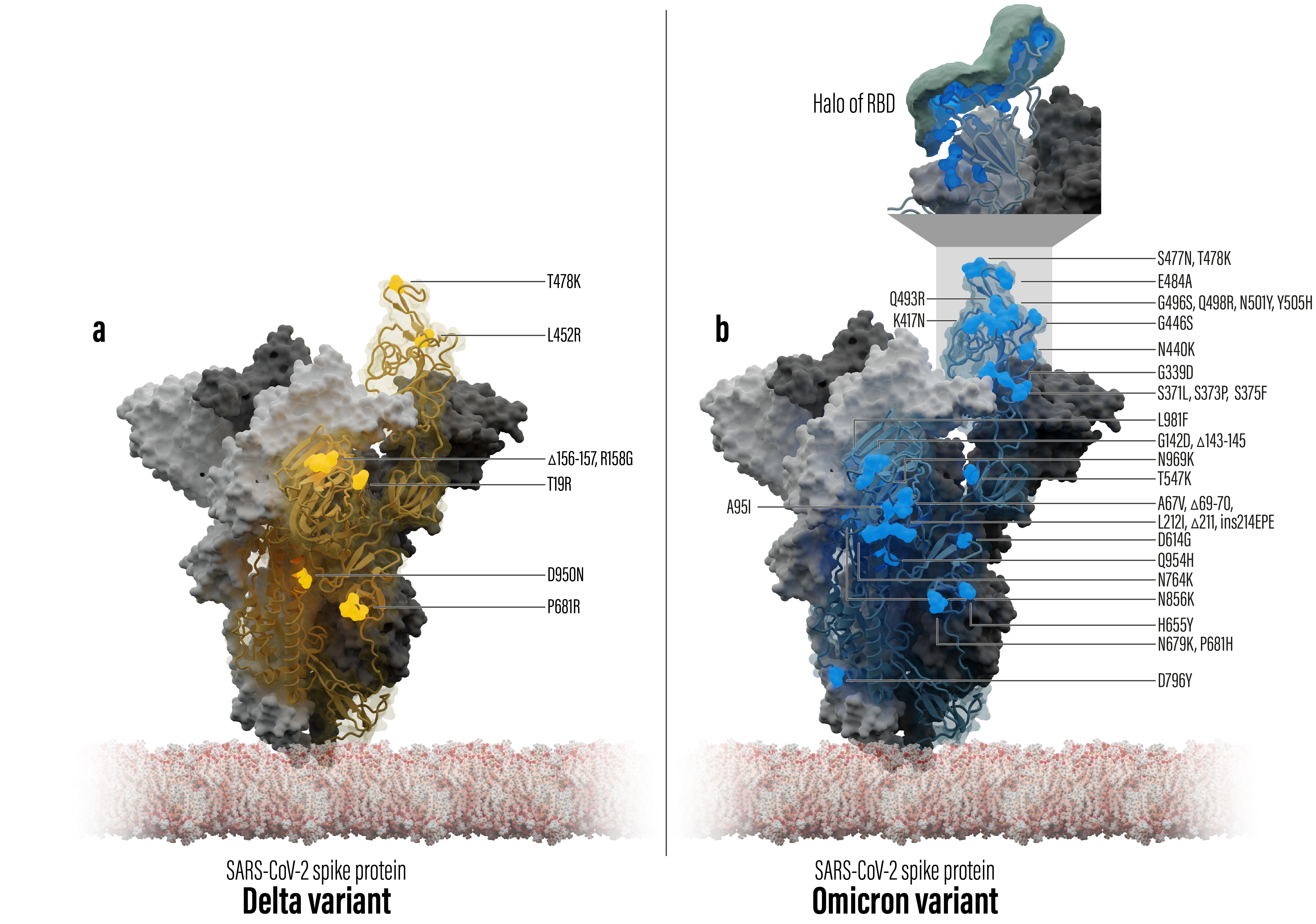
Fig 2: Comparison of mutation sites of Delta variant versus Omicron variant
Tracking the evolving virus over time (see video below), shows a high rate a of mutations arising with the Omicron variant. Spheres depict alpha-C-atoms of the corresponding amino acid residue. Both color and size correlate with the number of mutations at each position.
Phase 2: React
We are clearly in need of a structural bioinformatics analysis of this variant, in particular its expected binding mode and predicted affinity and this is what we set out to do.
To enable a comprehensive structural analysis of emerging partial or complete SARS-CoV-2 genomes, we follow a three-step approach.
Step 1: Analyze new genomes (e.g. from GISAID or sequences directly provided by associated laboratories or global and regional waste-water sampling) for sequences with amino acid exchanges in regions of SARS-CoV-2 protein structures.
Step 2: Look for sequences where one or more mutations are found that have not yet been investigated. Those sequences are submitted to a structure modeling workflow and are analyzed using our Catalophore Halo technology (illustrated in the video below showing a Halo depicting the surface environment in a detailed manner where each point represents a property given at a certain location).
Step 3: Compare any modified RBD’s Halo to the wild-type Halo generating a difference Halo. In case the Halo comparison shows a substantial change, we expose the new variant to an ESF-based molecular dynamics modeling pipeline to predict the corresponding change in binding affinity.
Phase 3: The outcome
Difference Halos help to track changes of physico-chemical properties when comparing surfaces of SARS-CoV-2 variants of concern RBDs in a qualitative manner and help to rapidly identify changes in RBD/hACE2 interface fields guiding a decision of which variants to simulate in full atomistic detail. Knowing that electrostatics or hydrophobicity may be associated with significant alterations in biomolecular interactions and the viral pathogenicity, this gives us a head start for newly emerging variants of concern.
Based on the linear interaction energy (LIE) method, we have developed an empirical binding affinity estimator that predicts pretty accurate binding energies for spike-RBD-hACE2 complexes at moderate wall-clock run times, especially using massive cloud computing facilities.
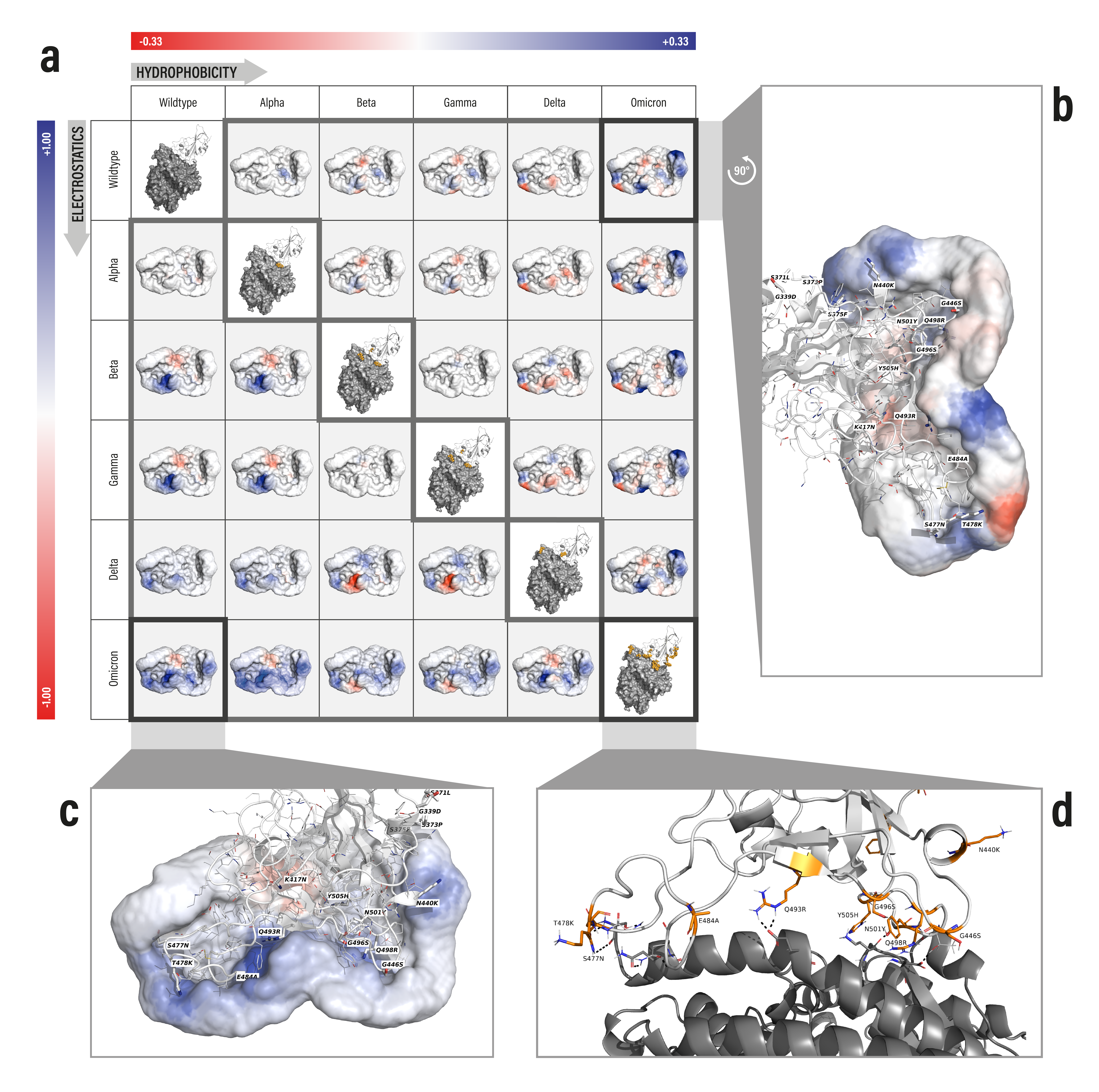
With the necessary caution, this technique helps to raise flags to indicate potential higher infectiousness. The precision of our SARS-CoV-2 spike-RBD-hACE2 binding affinity model stems from a large number of replicates and a solid and adequately large training set from one source.[13] Applying technology like this at scale enables the pharmaceutical industry and policy makers to have a head start on the evolution of a pandemic.
Cloud-powered diagnostic tools are critical in the fight against COVID-19 and other infectious diseases. Over the last year, AWS has seen inspirational results from the Diagnostic Development Initiative. We look forward to helping Innophore and other organizations worldwide use the cloud to mitigate current and future infectious disease outbreaks. Innophore's Catalophore technology is a great example of successfully scaling Big Data biotech applications in the AWS cloud. We are pleased to support their efforts to mitigate current and future infectious disease outbreaks.
Maggie Carter
Global Lead AWS Global Social Impact Team - California, USA
The future is: AI
Of course, our efforts do not end here. Bioinformatic predictions are only as good as they compare to real measurements.

© Innophore, March 2021
That's why our team specialists were trained to work in our partner BSL3 lab at the Medical University of Graz conducting measurements with live samples of SARS-CoV-2 variants and other pathogens in cell models.
This allowed us to further refine our pipelines and generate large amounts of data to train a robust AI model that predicts the affinity measures described here based on Catalophore halos in a fraction of the time. The availability of such a rapid model even enabled the optimization and design of novel therapeutics, which we describe in one of our recent manuscripts[14]. On-premise, predictions of a single virus variant require up to 2,000 CPU hours and five days each, heavily restricting processing at scale. Consequently, Innophore and AWS further collaborated to screen more than 14 million emerging virus genomes in the past 12 months. Applying this AI algorithm on AWS, calculations take 15 minutes per variant, bringing the total compute cost down by 95%.
So far, fortunately, the pandemic has lost most of its scare. To keep it that way, we need to keep a close eye on this virus and other biological threats[15]. We have learned that SARS-CoV-2 evades the immune system response through modifications of the spike protein, but only recently additional intense mutational dynamics have been observed worldwide: Since the global introduction of antiviral drugs targeting the main protease of SARS-CoV-2, a dramatic number of variants have been observed that alter the binding site for these drugs[16].
A technical blog post on amazon.com describes some details about running our simulations in the AWS cloud. Stay tuned for more Nature Health blog articles in this series, as our next papers are published soon.
Last edit: Sat Jan 14th, 2023
The people and resources behind this work
Coming back to the aforementioned flexibility: Most of the work was started and carried out during the start and the heights of the pandemic wave and was done remotely featuring different home offices in different countries. Challenges arising in this project were tackled by a diverse team of scientists. Molecular biologists, biotechnologists, (bio)chemists, physicists, machine-learning experts, and computer scientists teamed up to build a sturdy workflow for the evaluation of new variants of concern. The simulation performance was optimized in cooperation with Amazon Web Services (AWS), who supplied the necessary cloud infrastructure in the framework of the diagnostic development initiative.
About the companies behind the paper
Based in Austria, Innophore is a high-tech spin-off of the University of Graz and the Austrian Center of Industrial Biotechnology, specializing in the fields of digital drug discovery and enzyme search using 3D point clouds, AI and Deep Learning.
Amazon Web Services (AWS), Amazon.com's cloud computing service, is the world's largest cloud service provider. Since its launch in March 2020, the AWS Diagnostic Development Initiative has helped more than 85 organizations around the world, ranging from nonprofits and research institutions to startups and large businesses. AWS intends to distribute $12 million in support this year to organizations worldwide through this program.
Try a method behind this paper yourself
If you regularly work with protein structures, download Innophore's latest Schrödinger PyMOL plugin CavitOmiX for free and generate your own AI-predicted protein structures, using NVIDIA's BioNeMo, DeepMind's AlphaFold or Meta's ESMfold . Furthermore, Catalophore™ cavities can be calculated directly for molecules loaded in PyMOL. More information is also available at the official PyMOL wiki.
References
[1] Steinkellner, G. et al. Identification of promiscuous ene-reductase activity by mining structural databases using active site constellations. Nat Commun 5, 4150 (2014).
[2] Gorgulla, C. et al. A multi-pronged approach targeting SARS-CoV-2 proteins using ultra-large virtual screening. iScience Volume 24, issue 2, 102021 (2021)
[3] Singh, A. et al. Serine 477 plays a crucial role in the interaction of the SARS-CoV-2 spike protein with the human receptor ACE2. Sci Rep 11, 4320 (2021).
[4] Schrörs, B. et al. Large-scale analysis of SARS-CoV-2 spike-glycoprotein mutants demonstrates the need for continuous screening of virus isolates. PLOS ONE 16(9): e0249254 (2021).
[5] Parums, D. V. Revised World Health Organization (WHO) terminology for variants of concern and variants of interest of SARS-CoV-2. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 27, e933622–e933631 (2021).
[6] Thye, A.Y.-K. et al. Emerging SARS-CoV-2 variants of concern (VOCs): An impending global crisis. Biomedicines 9, 1303 (2021).
[7] Chen, C. et al. Computational prediction of the effect of amino acid changes on the binding affinity between SARS-CoV-2 spike RBD and human ACE2. Proc. Natl. Acad. Sci. 118, e2106480118 (2021).
[8] Augusto, G. et al. In vitro data suggest that Indian delta variant B.1.617 of SARS-CoV-2 escapes neutralization by both receptor affinity and immune evasion. Allergy 77, 111–117 (2022).
[9] Ozono, S. et al. SARS-CoV-2 D614G spike mutation increases entry efficiency with enhanced ACE2-binding affinity. Nat. Commun. 12, 848 (2021).
[10] Shu, Y. & McCauley, J. GISAID: Global initiative on sharing all influenza data—from vision to reality. Eurosurveillance 22, 30494 (2017).
[11] Implications of the emergence and spread of the SARS-CoV-2 B.1.1.529 variant of concern (Omicron) for the EU/EEA. 7 (2021).
[12] Hadfield, J. et al. Nextstrain: Real-time tracking of pathogen evolution. Bioinformatics 34, 4121–4123 (2018).
[13] Zahradník, J. et al. SARS-CoV-2 variant prediction and antiviral drug design are enabled by RBD in vitro evolution. Nat. Microbiol. 6, 1188–1198 (2021).
[14] Koechl, K. et al. Optimizing variant-specific therapeutic SARS-CoV-2 decoys using deep-learning-guided molecular dynamics simulations. Sci Rep 13 (2023)
[15] Parigger, L. et al. Preprint https://doi.org/10.21203/rs.3.rs-1693803/v1
[16] Parigger, L. et al. Recent changes in the mutational dynamics of the SARS-CoV-2 main protease substantiate the danger of emerging resistance to antiviral drugs. Frontiers in Medicine 9, (2022)
Follow the Topic
-
Scientific Reports

An open access journal publishing original research from across all areas of the natural sciences, psychology, medicine and engineering.
Related Collections
With Collections, you can get published faster and increase your visibility.
Obesity
Publishing Model: Hybrid
Deadline: Apr 24, 2026
Reproductive Health
Publishing Model: Hybrid
Deadline: Mar 30, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in