Insertion Sequence elements can promote bacterial adaptation in the short run but constrain it over longer time-scales
Published in Ecology & Evolution

In the late 1940s, Barbara McClintock discovered that “genetic entities” that induced chromosomal rearrangements could change an organism’s phenotype. This discovery took place only 4 years after Avery, McLeod and MacCarty’s paper on the role of DNA on bacteria transformation and just 4 years before Hershel and Chase established the role of DNA as the material basis of genetic information. Since then, many types of mobile genetic elements have been discovered including transposons, plasmids, insertion sequences (IS), and integrative conjugative elements. However, their roles in evolution are often unclear or contradictory. On the one hand, their effects on their hosts can be devastating, as in the case of the P-element in Drosophila. On the other hand, they can provide an evolutionary bounty, as seen in the domestication of RAG1-RAG2 transposase in the V(D)J recombination system.
In bacteria, IS elements produce genetic variation. Upon transposition, they can lead to gene inactivation, activation, or more subtle modifications of gene expression. In addition, they can mediate large chromosomal rearrangements including inversions, deletions, and duplications by recombination between homologous copies. However, their fitness effects and role in the dynamics of adaptation by natural selection have puzzled evolutionary biologists for decades. These questions led to the start of a fruitful collaboration between Dominique Schneider and Richard Lenski, the founder of the well-known Long-Term Evolution Experiment (LTEE) with Escherichia coli (Figure 1). More than 20 years ago, they published their first collaborative work, and it was a study of IS-mediated changes during the LTEE. In this work, they characterized the molecular events that produced multiple IS-mediated mutations in two of the 12 LTEE lines. However, whether these mutations were beneficial and thus contributed to adaptation remained unclear1. It was only several years later that it was established that at least three of these mutations accounted for more than half of the fitness gain of an Ara+1 clone isolated at 2000 generations2-4. But what about the fitness effects of IS-mediated mutations over much longer evolutionary time-scales? It took more than 50,000 generations of the LTEE and over 23 years of the Schneider-Lenski collaboration to answer this question.

commemorative agar plate made for the 25th anniversary of the LTEE.
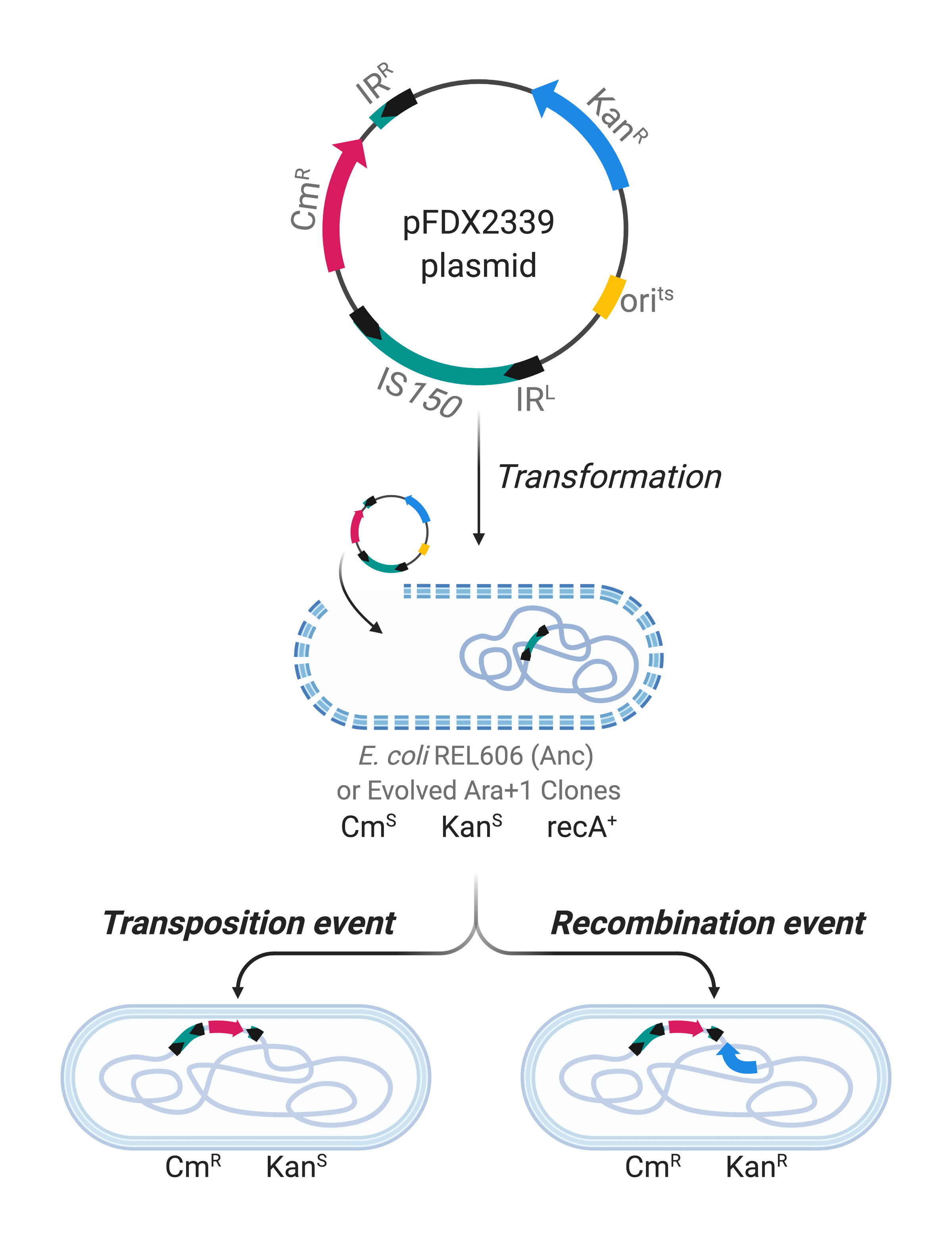
In our recent work published in Nature Communications, we used genomic and metagenomic data5,6, to characterize the distribution and analyze the contribution to genome evolution of IS elements in all 12 populations of the LTEE over 50,000 generations. We found that IS-mediated mutations accumulated significantly more slowly in populations that had evolved point-mutation hypermutability. Meanwhile, those hypermutable lines reached somewhat higher fitness levels than the populations that retained the low ancestral rate of point mutation7. These findings imply that the faster adaptation enabled by a greater supply of point mutations counteracts the successful invasion of beneficial mutations mediated by IS elements, and it also suggests that IS-mediated mutations may preclude some other genetic paths of adaptation. We further analyzed one population, called Ara+1, that both accumulated the most IS-related mutations (usually mediated by IS150) and had the smallest fitness gain over 50,000 generations. This population’s early burst of IS150-mediated mutations was not explained by mutations in any copies of the IS150 element itself. Using the pFDX2339 reporter plasmid8, we measured the rates of IS150 transposition from the plasmid into the chromosome, and of recombination between the plasmid-borne IS150 copy and copies of that element in the chromosome (Figure 2). These events increased early in this population’s history, but then later declined, a dynamic that is reminiscent of the emergence and decline of point-mutation hypermutability in other LTEE populations9. Altogether, our results show that IS elements can promote adaptive evolution in the short run, but over longer timescales they may constrain evolution by imposing a load of deleterious mutations and perhaps also closing off some other pathways to adaptation.
Open access to the paper here:
Consuegra, J., Gaffé, J., Lenski, R.E. et al. Insertion-sequence-mediated mutations both promote and constrain evolvability during a long-term experiment with bacteria. Nat Commun 12, 980 (2021). https://doi.org/10.1038/s41467-021-21210-7
References
- Papadopoulos D. et al. Genomic evolution during a 10,000-generation experiment with bacteria. PNAS. 1999). Doi: 10.1073/pnas.96.7.3807
- Philippe, N. et al. Evolution of penicillin-binding protein 2 concentration and cell shape during a long-term experiment with Escherichia coli. J. Bacteriol. 191, 909–921 (2009). Doi: 10.1128/JB.01419-08
- Barrick, J. E. & Lenski, R. E. Genome-wide mutational diversity in an evolving population of Escherichia coli. Cold Spring Harb. Symp. Quant. Biol. 74, 119–129 (2009). Doi: 10.1101/sqb.2009.74.018
- Woods, R. et al. Tests of parallel molecular evolution in a long-term experiment with Escherichia coli. PNAS 103, 9107–9112 (2006). Doi: 10.1073/pnas.0602917103
- Tenaillon, O. et al. Tempo and mode of genome evolution in a 50,000-generation experiment. Nature 536, 165–170 (2016). Doi: 10.1038/nature18959
- Good, B. H. et al. The dynamics of molecular evolution over 60,000 generations. Nature 551, 45–50 (2017). Doi: 10.1038/nature24287
- Wiser, M. J. et al. Long-term dynamics of adaptation in asexual populations. Science 342, 1364-1367 (2013). Doi: 10.1126/science.1243357
- Haas, M. & Rak, B. Escherichia coli insertion sequence IS150: transposition via circular and linear intermediates. J. Bacteriol. 184, 5833–5841 (2002). Doi: 10.1128/JB.184.21.5833-5841.2002
- Wielgoss, S. et al. Mutation rate dynamics in a bacterial population balance evolvability and genetic load. PNAS 110, 222-227 (2013). Doi:
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in