It’s time to spike
Published in Computational Sciences and Mathematics
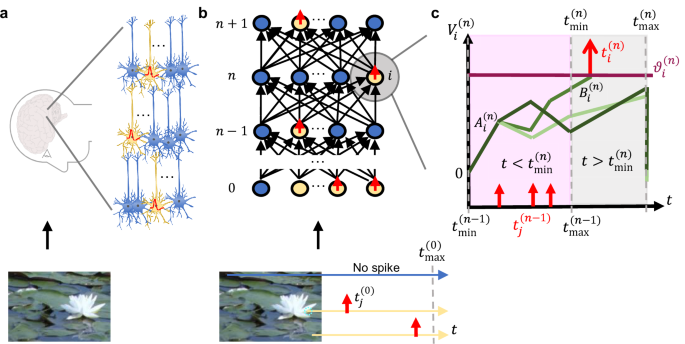
While the capabilities of deep-learning neural networks are impressive, this comes at a cost of huge amounts of consumed energy. In contrast, biological brains, which comprise neurons transmitting information via sparse voltage spikes, are highly energy efficient. Inspired by the energy efficiency of the brain, researchers have been exploring biologically plausible models of spiking neural networks (SNNs) that exploit spike timings for communication. The figure below schematically illustrates this concept, where exact rich information on image intensity can be encoded into the timing of sparse all-or-none spikes (the higher the intensity, the earlier the spike) that subsequently propagate through the network.
 Astonishingly, despite theoretical results enabling mapping of weights from ANNs to temporally coded SNNs, the training of SNNs to high accuracy has remained a challenge. We kept wondering, “Why is the accuracy gap still there?”, and decided to track the root cause down to the tiniest detail of training.
Astonishingly, despite theoretical results enabling mapping of weights from ANNs to temporally coded SNNs, the training of SNNs to high accuracy has remained a challenge. We kept wondering, “Why is the accuracy gap still there?”, and decided to track the root cause down to the tiniest detail of training.
As reported in Nature Communications, we have analyzed in theory and simulation the learning dynamics of time-to-first-spike SNNs and identified a specific instance of the vanishing-or-exploding gradient problem. To close the accuracy gap between the ANNs and the SNNs, we proposed an approach for SNN training that is characterized by ANN-equivalent, or more specifically ReLU-equivalent, training trajectories.
In search for training instabilities
From a practical perspective, even the most powerful and efficient model is of limited use, if it cannot be tuned well. It took quite some time for the ANN research to establish effective training procedures that enabled reliable training. Aspects, such as proper initializations, proper optimizers and numerically stable network formulations, are results of many years of research. We realized that similar effort has been missing for SNNs, where researchers often directly reuse the training solutions developed for ANNs.
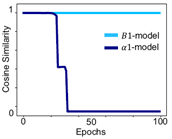
We hypothesized that this might be hindering SNN accuracy, and our intuition has proven right. Indeed, we found a vanishing-or-exploding gradient problem in SNNs during training – a problem also historically present in early ANNs. One of the solutions for this problem is specific weight initialization. In our work, we introduce a “smart initialization” for SNNs that ensures gradient stability. Yet only initially… After a certain number of training epochs, the weights are attracted again towards unstable regimes. We realized that there is a systematic training trajectory bias, which motivated us to analyze the details of the SNN model formulation and their impact on the training trajectory. This brings us to the key contribution of our work: we propose a specific SNN formulation that we call the B1 model. The figure below illustrates the alignment of training trajectories (measured as cosine similarity: the closer to one, the better) between the conventional ReLU ANN model and: 1) the prior art SNN α1 model with our smart initialization, and 2) our SNN B1 model. Our model ensures that during the training the SNN follows equivalent training trajectories to a ReLU-based ANN for the entire training, which is visible in the figure as maintaining cosine similarity equal to one throughout the entire training. In consequence, our approach can be effectively used to train SNNs to ANN-equivalent accuracy.
Benchmarking results and their impact
We demonstrated ANN-equivalent classification accuracy, surpassing previous state-of-the-art SNN models, on datasets such as MNIST, Fashion-MNIST, CIFAR10, CIFAR100 and PLACES365. We also measured the average number of spikes required per neuron per each inference example. Our approach accomplishes high-performance classification with less than 0.3 spikes per neuron, lending itself to an energy-efficient implementation.
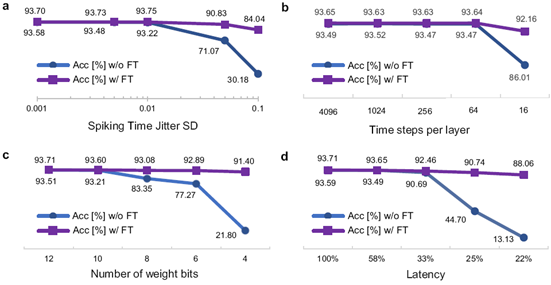
Furthermore, the capability to train SNNs directly enables fine-tuning the models to unlock the full potential of neuromorphic hardware. We simulated various hardware characteristics: random noise imposed on spiking times, operation with constrained temporal resolution, number of weight bits, and maximum permitted latency. The figure below illustrates that the accuracy with fine-tuning (w/ FT), enabled by our approach, largely recovers or even surpasses the accuracy of the models without fine-tuning (w/o FT).
 In the future, our theory could also serve as a starting point to develop stable training approaches for other SNN models with temporal coding, and to devise learning rules directly implementable in hardware. We envision that a promising use case would involve operation on devices undergoing continual online learning on the chip, ensuring energy-efficient and low-latency inference. For further technical details, please refer to our open access research paper “High-performance deep spiking neural networks with 0.3 spikes per neuron.“
In the future, our theory could also serve as a starting point to develop stable training approaches for other SNN models with temporal coding, and to devise learning rules directly implementable in hardware. We envision that a promising use case would involve operation on devices undergoing continual online learning on the chip, ensuring energy-efficient and low-latency inference. For further technical details, please refer to our open access research paper “High-performance deep spiking neural networks with 0.3 spikes per neuron.“
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in
ANN is not yet at its matured stage.
Looking forward