Jurassic Park in the chemical world – reconstructing ancient proteins to design optimal biocatalysts for the future.
Published in Chemistry
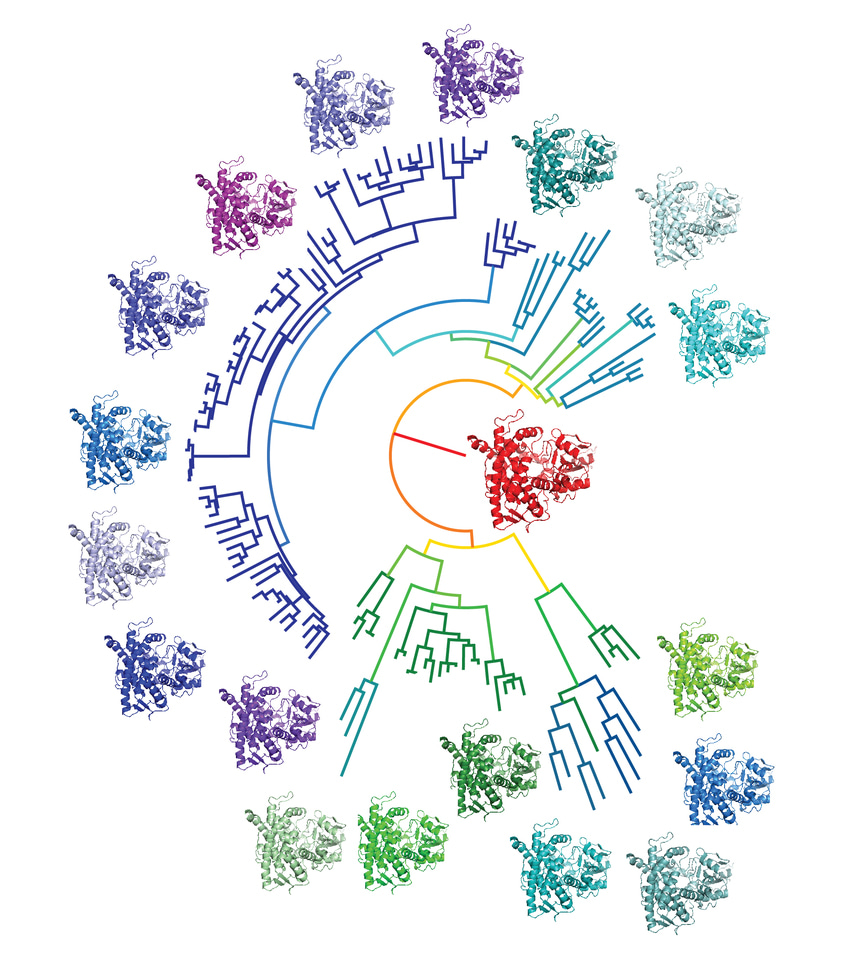
Enzymes are highly efficient catalysts that facilitate all chemical reactions essential in living organisms. Without enzymes, life as we know it on our planet would not have evolved. Due to their catalytic potential, enzymes have also increasingly been incorporated into commercial processes for the production of high-value chemicals. However, enzymes evolved to work under physiological, not industrial, conditions, so a serious limitation for the use of enzymes as industrial biocatalysts is their limited stability, especially at the elevated temperatures and over the extended incubation times frequently encountered in industrial bioreactors. While directed evolution has been used to improve the stability of enzymes, this approach is generally time-consuming and may require the screening of a large number of variants to identify the most suitable candidate catalysts. A simpler and more direct alternative might be to delve into the evolutionary history of enzymes and resurrect an ancestor.
Recently, a method has emerged that allows the sequence of an ancestral precursor of extant enzymes to be inferred from the sequences of modern relatives1. Ancestral Sequence Reconstruction (ASR) is a real-world enactment of Michael Crichton’s Jurassic Park on the molecular level – instead of creating rampant dinosaurs in the laboratory, this method allows us to make ancient enzymes that may have existed eons ago. However, typically this involves going much farther back than to the dinosaurs, usually to the time when the environment on the Earth was much hotter than today and all life was marine and unicellular. It has already been demonstrated2 that this method can lead to enzyme variants with improved thermostability - enzymes that worked under conditions present on the early Earth had to be stable at temperatures around 60°C or more. In the scenario created on Jurassic Park’s Isla Nublar, DNA found in some blood droplets of a well-preserved mosquito enabled scientists to create organisms that roamed the planet a long time ago. In contrast, ASR predicts DNA sequences (and corresponding protein sequences) using bioinformatics algorithms. While this method only allows us to infer the most probable ancestor, not necessarily the historically correct one, it still provides an exciting platform from which to recreate history for beneficial purposes.
We used the cytochrome P450 enzymes as a model enzyme3. P450s are a very large group of haem-dependent monooxygenases that catalyze regio- and stereoselective functionalizations of unactivated C-H bonds, a type of reaction that is challenging to perform using purely chemical methods. Surprisingly we only had to go back ~ 450 million years (to the very first vertebrates) to find an ancestor that could tolerate temperatures around 60°C or more, but which retained the useful catalytic activities of the extant enzymes from humans. It was also relatively tolerant of solvents, another very useful property for industry. To demonstrate the generality of this method, we tested it on another unrelated enzyme, ketol-acid reductoisomerase (KARI). KARI plays an essential step in the synthesis of branched-chain amino acids in plants and microorganisms and can be used in synthetic enzymatic cascades for the production of biofuels. Here, a considerable improvement in catalytic efficiency was observed, as well as improved thermostability.
This approach should make it possible to devise thermostable biocatalysts much more quickly and efficiently and use them as templates for further evolution in an approach we call CLADE: Combinatorial Libraries of Ancestors for Directed Evolution. This promises to open up new opportunities for using proteins in synthetic biology. However it might also help in shedding light on the ‘dark proteome’ - it is much easier to study and structurally characterize proteins that are stable. Ultimately, this will lead to a better understanding of how Nature has shaped these powerful biocatalysts to do useful biological chemistry and how we can repurpose them to make industrial chemistry cleaner, greener and more sustainable.
Written by Elizabeth Gillam and Gary Schenk
References
1 Cole, M. F. & Gaucher, E. A. Utilizing natural diversity to evolve protein function: applications towards thermostability. Curr. Opin. Chem. Biol. 15, 399-406, doi:10.1016/j.cbpa.2011.03.005 (2011).
2 Gaucher, E. A., Thomson, J. M., Burgan, M. F. & Benner, S. A. Inferring the palaeoenvironment of ancient bacteria on the basis of resurrected proteins. Nature 425, 285-288, doi:Doi 10.1038/Nature01977 (2003).
3 Gumulya, Y. et al. Engineering highly functional thermostable proteins using ancestral sequence reconstruction. Nat. Catal., doi:10.1038/s41929-018-0159-5 (2018).




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in