Label-free photoacoustic histology for intraoperative diagnosis of bone
Published in Bioengineering & Biotechnology
Despite the rapid advances in cancer treatment, surgical excision of tumors is still preferred for most cancer patients if possible. In oncology surgery, intraoperative pathological diagnosis is effective and essential, which provides surgical guidance and identification of tumor margins and helps patients to avoid second surgeries. Oncologic surgeons currently rely on the frozen section technique to provide rapid pathological examination and guide tumor resection, which usually suffers from the freezing artifact with compromised imaging qualities and is difficult for borderline tumors.
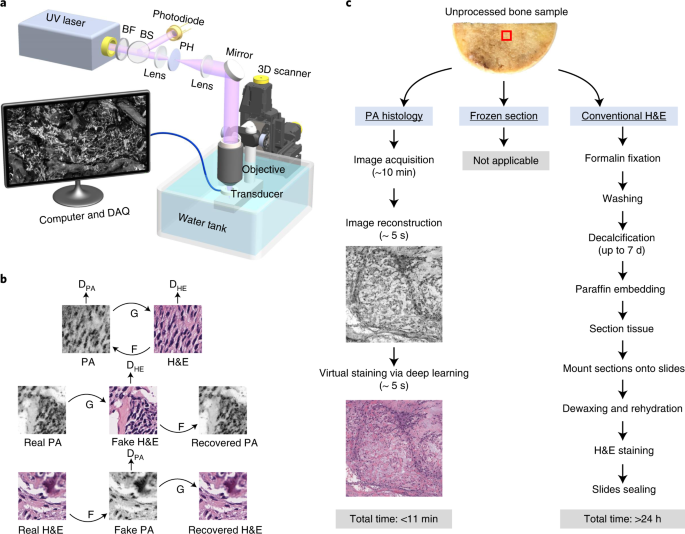
To fill this gap, we developed label-free ultraviolet photoacoustic microscopy (UV-PAM) for fast histology-like imaging back in 20171. That UV-PAM system utilizes optical absorption-induced ultrasound signals to acquire histology-like images of sectioned tissue slices. The label-free PAM images showed high correlations to the conventional histologic images and are reviewed by pathologists to identify tumors. Without the need for staining and sectioning, the technique has the potential to be a powerful tool for intraoperative pathological diagnosis. However, the previous system was built in transmission mode, limiting its applications to only thin slices and thus preventing it from rapid slide-free histology. Later, we developed a microtomy-assisted UV-PAM for thick samples and demonstrated 3D imaging of whole organs, which used a microtome to cut the top of agarose-embedded tissues into a flat surface for high-resolution imaging2.
When we discussed potential histology applications with surgeons and pathologists, we realized the time cost of sample preparation procedures (i.e., agarose embedding and cutting) is not ideal for intraoperative use. The ideal intraoperative imaging system should require minimal sample preparations, such as cutting, embedding, freezing, or staining. However, the excised tissues usually have irregular surfaces without further cutting or squeezing, making it challenging for high-resolution imaging. Although z-stack imaging, which scans at different z-positions, can be used to acquire high-quality images of uneven surfaces, it significantly increases the imaging time and thus is not practical for intraoperative use. Fortunately, the ultrasound time-of-flight information can be used to measure the axial position with high accuracy in UV-PAM, allowing us to dynamically adjust the z-position of our specimen and ensure it is located within the depth of field. Based on this, we developed the three-dimensional contour scanning UV-PAM system to image slide-free specimens for histology-like images.
Meanwhile, Dr. Brooke Crawford, an orthopaedic oncology surgeon who routinely removes tumors from the musculoskeletal system, approached our lab and mentioned the long-standing challenge of intraoperative bone histology. To slice the bone tissue thin enough to create a microscope slide requires “softening” the bone by removing the calcium; this can take several days. So, unlike soft tissue tumors, where she can look at the edges of the tumor or margin while the patient is still under anesthesia, bone tumors require her to use measurements taken from imaging done before the surgery – and she usually adds 2 centimeters beyond the tumor to the measurement before cutting the tissue. This is the only way she and other orthopaedic oncology surgeons can be sure about the removal of the whole tumor. But 2 cm is a lot, especially since surgeons must add it in all directions of the bony portion of the tumor. It can mean the difference between saving a joint, saving enough bone to fix instead of replacing it, and determining the way to reconstruct the bone. Then, we realized that the UV-PAM could be used to look at unprocessed bone specimens quickly and accurately for intraoperative diagnosis, which may have a big impact on the clinic.
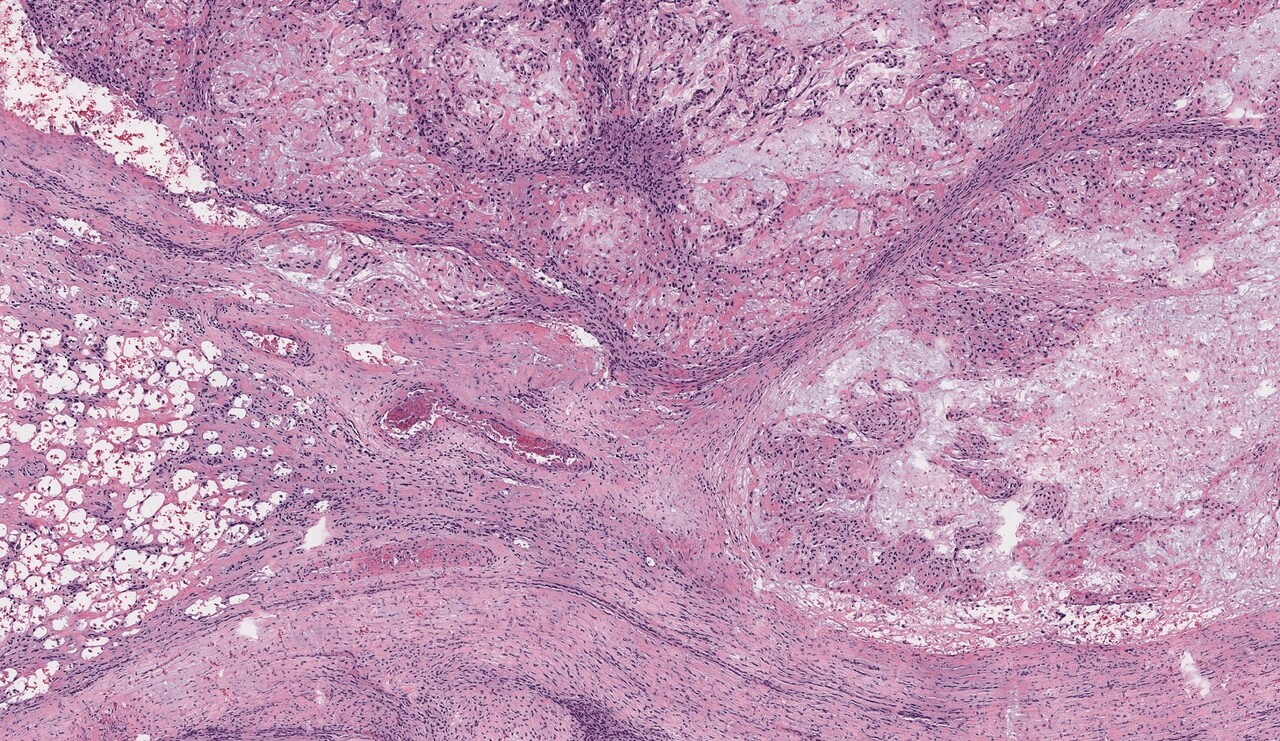
After we got some histology-like images of bone specimens, we showed the results to several pathologists and asked them if they could make diagnostic decisions using our grayscale UV-PAM images. Although they can identify the tumor and differentiate normal areas and tumor areas in the grayscale UV-PAM images, the color difference makes this process not as easy as conventional H&E images since the pathologists are used to the H&E color. Thus, we decided to use deep learning techniques to virtually stain our images to better match the conventional H&E slides, which will be more interpretable for pathologists. For this purpose, we chose the cycle-consistent generative adversarial network (i.e., CycleGAN) for unsupervised deep learning, which has shown superior performance in image processing and color conversion3. With this virtual staining technique and pre-trained neural network, we can generate H&E-like images for rapid intraoperative pathological diagnosis (Figure. 1) within minutes.
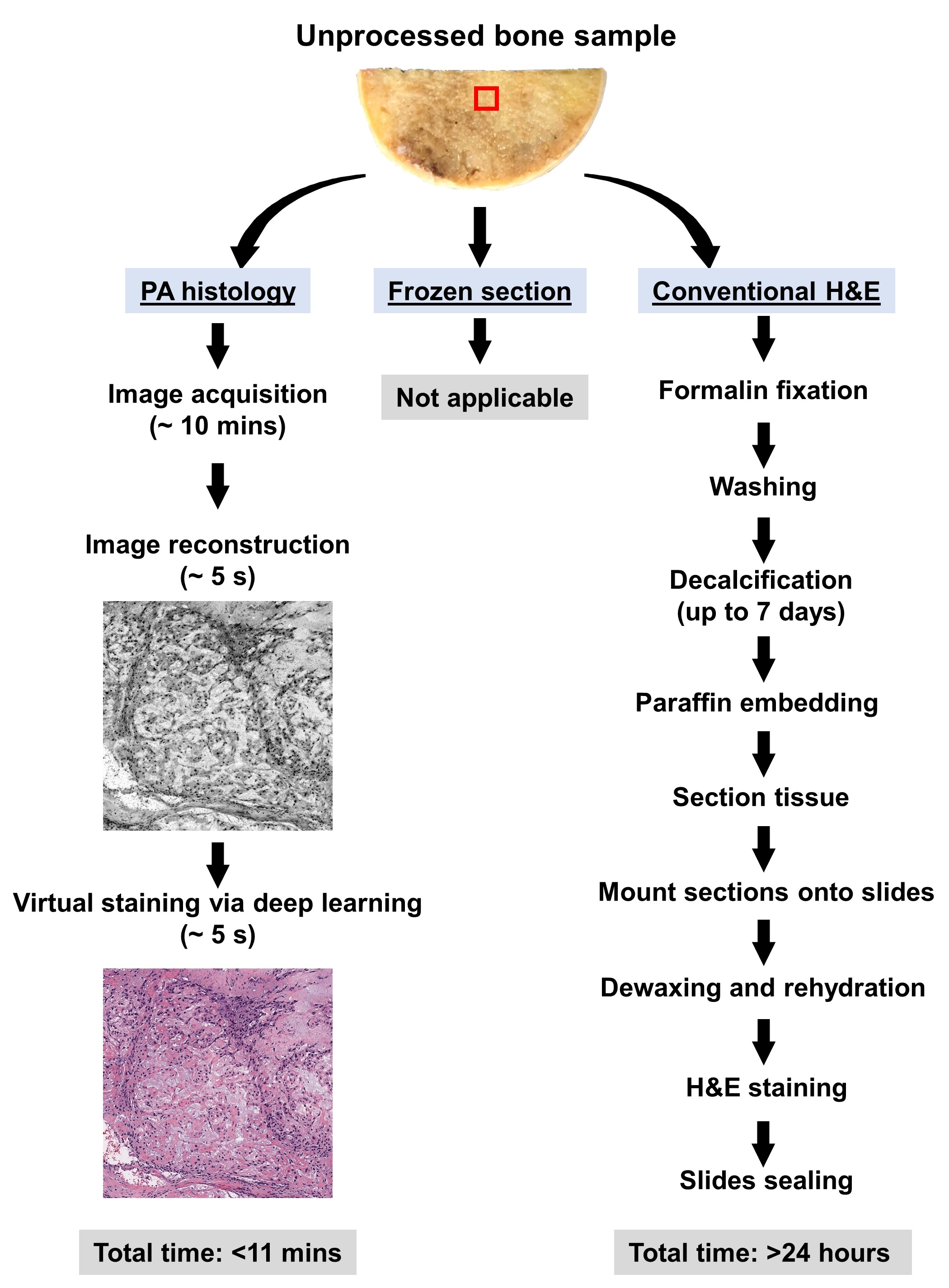
Figure 1. The workflow for PA histology and conventional H&E staining histology of bone samples.
We have presented our results to the orthopaedic community during major conferences and got incredibly exciting feedback from orthopaedic surgeons and pathologists. Outside of Dr. Crawford’s specialty, many of her orthopaedic partners are very interested in this technology. Although we used bone tumors to test the capabilities of photoacoustic microscopy, using it to check the margins of tumors is only one of many implications of this tool. It could also be used for benign bone tumor removals, which involve scraping the tumor out of the bone, and the completeness of the scraping can determine whether the tumor returns. With future development, it could be used to monitor metabolic bone conditions like osteoporosis, identify bone infections, and monitor fracture healing. Our next big step is setting up a system at a hospital so there is easy access to specimens and pathologists so we can fulfill the great potential of this technology. We hope that we can develop a hand-held probe to run over the scraped cavity and know if any further scraping is needed. In addition, we will continue to utilize deep learning techniques to provide a real-time diagnosis to assist surgeons and pathologists.
These results were recently published in Nature Biomedical Engineering: R. Cao, S. D. Nelson, S. Davis, Y. Liang, Y. Luo, Y. Zhang, B. Crawford*, and L. V. Wang*, Label-free ultraviolet photoacoustic histology via deep learning for rapid intraoperative diagnosis of bone cancer, Nature Biomedical Engineering, 2022, 10.1038/s41551-022-00940-z.
References:
- Wong, T. T. W. et al. Fast label-free multilayered histology-like imaging of human breast cancer by photoacoustic microscopy. Science Advances 3, e1602168 (2017).
- Wong, T. T. W. et al. Label-free automated three-dimensional imaging of whole organs by microtomy-assisted photoacoustic microscopy. Nat Commun 8, 1386 (2017).
- Zhu, J.-Y., Park, T., Isola, P. & Efros, A. A. Unpaired Image-to-Image Translation Using Cycle-Consistent Adversarial Networks. in 2017 IEEE International Conference on Computer Vision (ICCV) 2242–2251 (IEEE, 2017). doi:10.1109/ICCV.2017.244.
Follow the Topic
-
Nature Biomedical Engineering

This journal aspires to become the most prominent publishing venue in biomedical engineering by bringing together the most important advances in the discipline, enhancing their visibility, and providing overviews of the state of the art in each field.


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in