LEVA: Patterning Extracellular Vesicles and Particles with Light
Published in Bioengineering & Biotechnology

Explore the Research
Light-induced extracellular vesicle and particle adsorption - Nature Methods
LEVA is a label-free immobilization method for studying surface-bound extracellular vesicles.
What if we could arrange single extracellular vesicles (EVs), like pixels on a screen, to draw complex shapes and designs? This question sparked the development of Light-Induced Extracellular Vesicle and Particle Adsorption (LEVA)—a method born of curiosity to understand the role of EVs in living systems, collaboration, and serendipity.
Understanding EV Individuality
EVs are lipid nanoparticles released by cells that act like nanoscale postal packages carrying messages that influence the behavior of recipient cells. What makes EVs particularly difficult to study is their high diversity, which varies across message content, package size, and delivery method. Conventional methods measure these characteristics in bulk, obscuring the diverse content of individual particles.
Our team aimed to decipher these messages by tethering single EVs onto glass surfaces with antibodies and detecting their molecular cargo with high-resolution fluorescence microscopy. While the method successfully sorts and detects single EVs, we wanted a technology that could bridge molecular quantification with cell functional assays. Inspired by migrasomes, matrix-bound EVs left as “breadcrumbs” by migratory cells, we envisioned creating synthetic EV trails using surface chemistry approaches while preserving our single-EV resolution.
A Collaborative Journey
To produce these synthetic EV trails, we assembled around the drawing board. Each researcher contributed their unique expertise, ranging from EV biomanufacturing, photochemistry, neutrophil biology, bacterial engineering, and computational modeling. We had previously used UV photopatterning to generate protein micropatterns for cell functional assays and antibody-based EV biosensors, leading us to ask: since EVs are decorated with proteins, can we circumvent antibody-based tethering by leveraging the adsorption properties of proteins on the EV surface?
Sure enough, the EVs bound to the micropatterned surface within minutes.
Applications and Surprises
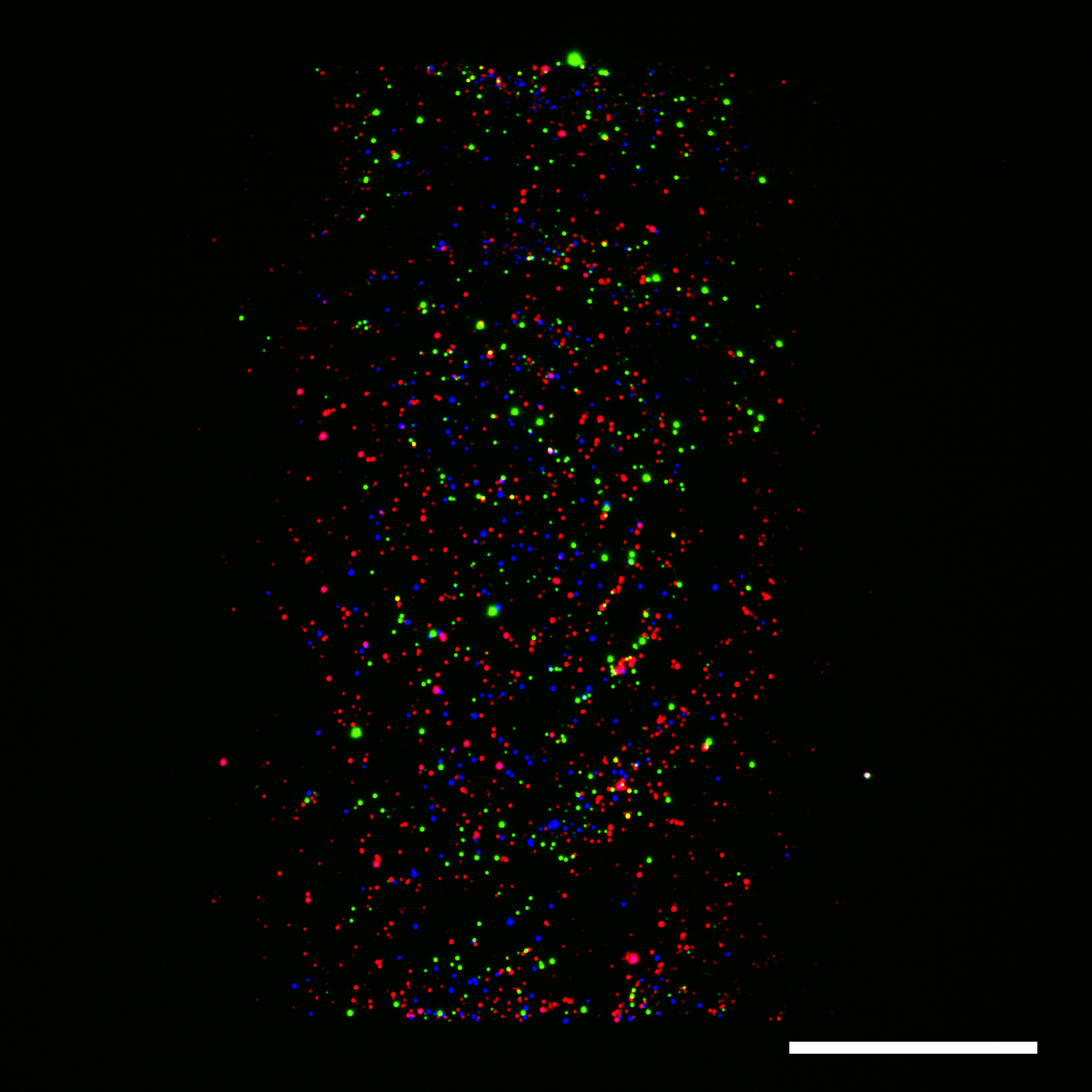
Having a method that precisely immobilizes EVs, we set a goal to pattern every EV under the sun. We patterned different-sized EVs, EVs from various taxonomic kingdoms, commercial EVs, and newly discovered non-vesicular extracellular particles known as exomeres. All extracellular particles patterned successfully. LEVA was ready for applications.
Our first application investigated whether LEVA could distinguish EV sizes through binding kinetics. Indeed, small EVs bound faster than large EVs. Our computational model, which assumed electrostatic interactions as the primary driving force for binding, mirrored our experimental results. This finding revealed that the negative charge on the EV surface and the positive charge on the micropattern also contributed to EV binding beyond adsorption.
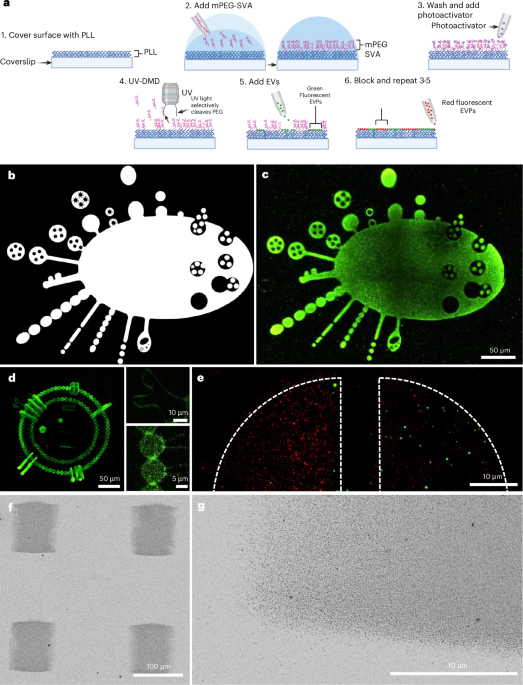
Unlike most micropatterning techniques, which are binary—either patterned or not—photopatterning can produce protein gradients by modulating UV light intensity. For our second application, we tested this ability to titrate EVs digitally. Similar to proteins, EVs adsorbed according to the pre-designed gradients. To answer our original question, we pushed the boundaries of this capability by creating grayscale images with single EVs by tuning EV density, similar to adjusting pixel density. My favorite EV drawing was an EV made entirely of single EVs, which we call “EVception” (shown in Fig. 1d in the manuscript).
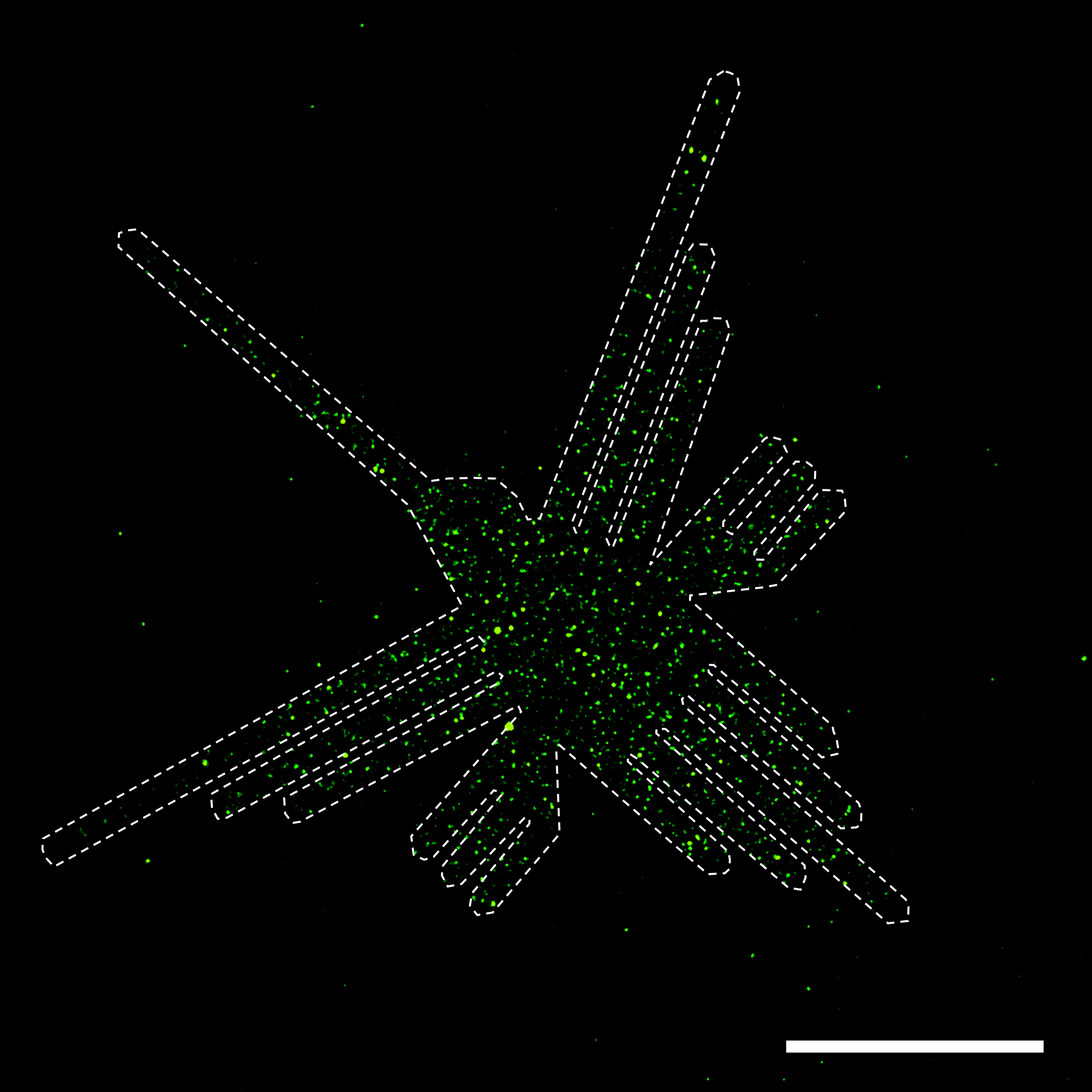
We had demonstrated LEVA's tunability, but still needed to show its ability for cell functional assays. To generate fluorescent EVs, we engineered green fluorescent protein (GFP)-expressing bacteria that coincidentally produced GFP-positive EVs. The next question was: can we use these EVs to elicit a cellular response? Our lab develops bacterial patterns to study neutrophil swarming, our body’s first line of defense against foreign invaders and tissue damage. Given our ability to produce large-scale arrays of bacterial EVs resembling our bacterial patterns, we added neutrophils, unsure whether they would swarm around the bacterial EVs. Without much suspense, the neutrophils swarmed around the EV patterns, just as they did with our bacterial patterns, but with the additional benefit of producing complex swarming patterns in a matter of minutes, which is arduous with our bacterial method.
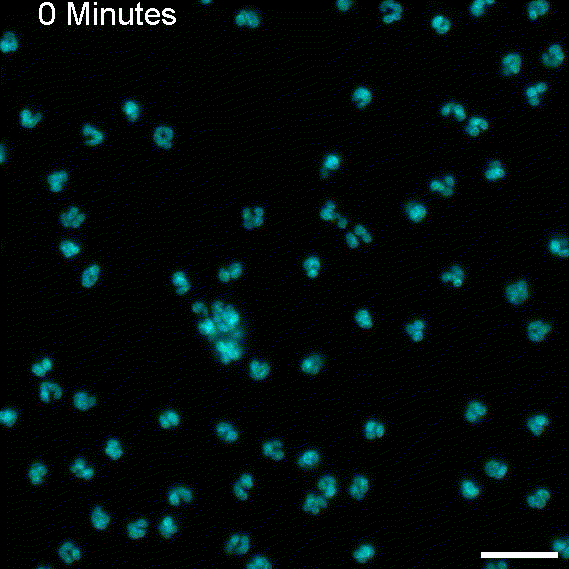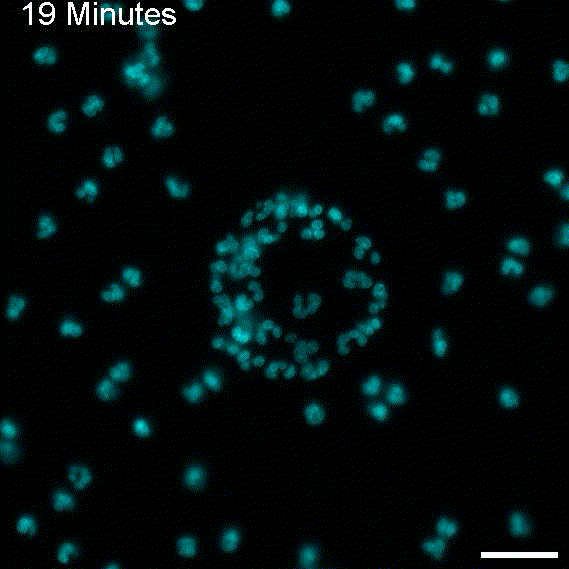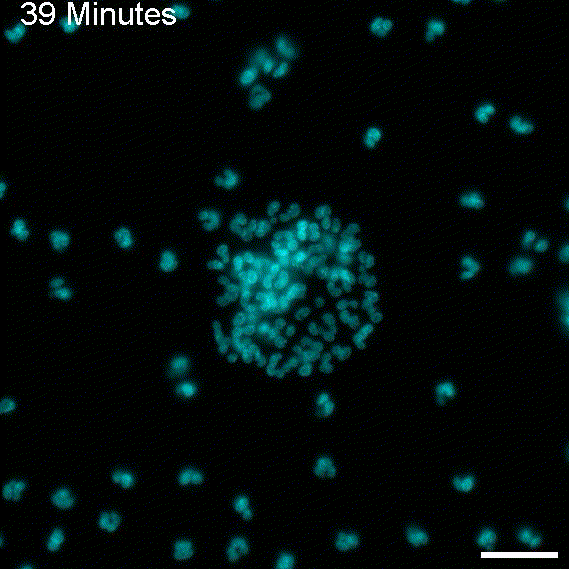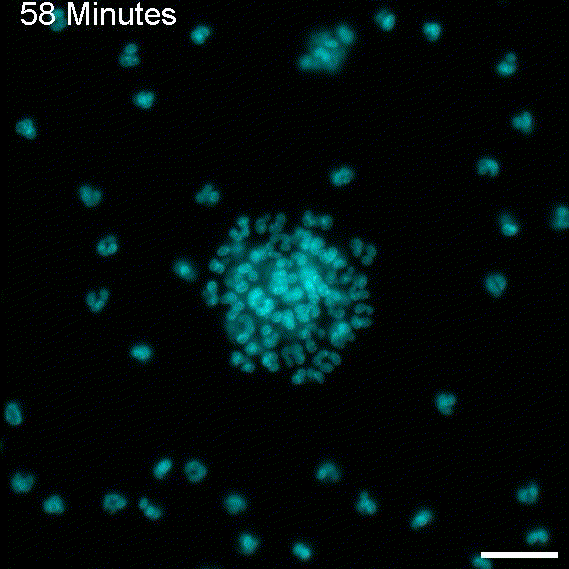
To return this application to its original inspiration, we created large EV tracks and added glioblastoma cells. As expected, the cells followed the tracks, as observed with migrasomes.
Looking Ahead
Developing LEVA was one of those rare moments that occurred in the right place, at the right time, with the right people. What began as a simple question has evolved into a tool that paves the way for new avenues and paths yet to be explored, offering fresh perspectives on EVs and their role in health. We are excited to see how others utilize LEVA, whether to decrypt messages sent between cells, organs, and organisms, or to create molecular art. For us, the journey is far from over, and we are excited to continue using LEVA to unlock the secrets of EV biology.
Follow the Topic
-
Nature Methods

This journal is a forum for the publication of novel methods and significant improvements to tried-and-tested basic research techniques in the life sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Methods development in Cryo-ET and in situ structural determination
Publishing Model: Hybrid
Deadline: Jul 28, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in