Life’s ‘original sin’ allowed the emergence programmed cell death in the earliest cellular organisms
Published in Ecology & Evolution

Programmed cell death (PCD), “an active, genetically controlled, cellular self-destruction driven by a series of complex biochemical events and specialized cellular machinery” (Berman-Frank et al. 2004), played an important role in the evolution of life. For example, PCD was essential for the major evolutionary transitions in individuality, such as eukaryogenesis, multicellularity, and eusociality in insects (Iranzo et al. 2014; Durand, Barreto Filho and Michod 2019; Blackstone and Green 1999). What is controversial, however, is how and why PCD originated at all in prokaryotes.
PCD was initially considered a unique feature of multicellular life, and studies of cell death were focused primarily on eukaryotes. However, the discovery of PCD in bacteria and the accumulating evidence shows that PCD is universal in unicellular and multicellular life (Engelberg-Kulka et al. 2006; Allocati et al. 2015; Peeters and de Jonge 2018). Taken together, amounting evidence suggests that PCD may have a much more ancient origin than previously thought. Discussions of prokaryote PCD were dominated by bacteria, and archaea have almost been entirely excluded due to the limited genomic data available (Bidle et al. 2010). For that reason, I wished to investigate the essential members of the PCD genetic toolkit, caspases (CAs) and their homologs, in the archaea and examine their evolutionary history.
Although other CA-independent PCD pathways exist, CAs and their homologs are frequently used to study the evolution of eukaryotic PCD (Klim et al. 2018; Koonin and Aravind 2002). It is important to note that various members of CA homologs exist, including metacaspases (MCAs), paracaspases (PCAs) and orthocaspases (OCAs) (Minina et al. 2020). They are all classified within the C14 peptidase family (Rawlings et al. 2014). In addition, CAs and CA homologs have different domain architectures and are associated with different functions that include both pro-life and pro-death functions (Minina et al. 2020).
Identification and classification of CA homolog of sequences in public databases revealed OCAs and type I MCAs are surprisingly widespread in archaea and across the tree of life (Figure 1). In line with earlier predictions by Choi and Berges 2013, OCAs were the most ancestral members because of their presence in the deepest branching archaeal lineages. These OCAS exhibited conservation of key residues and structural similarities with eukaryotic CAs, PCAs and MCAs. This led me to my next research question: are these PCD effectors ancestral to both archaea and bacteria or did they arise after the divergence of the two prokaryotic lineages with subsequent horizontal gene transfer (HGT)?
Subsequent phylogenetic reconstruction analyses of OCAs and type I MCAs across the three domains of life revealed incongruency between gene and species trees, which is most likely due to extensive HGT between archaea and bacteria. This is expected as large-scale HGT events were common between proto-cellular organisms during the emergence of diverse prokaryotic lineages (Koonin 2009). While alternative explanations may exist, the incongruency between species and gene trees as well as the wide taxonomic distribution in all three domains of life suggest that OCAs predate the divergence of archaea and bacteria, possibly as far back as LUCAS (Last Universal Common Ancestor State) (Figure 1).
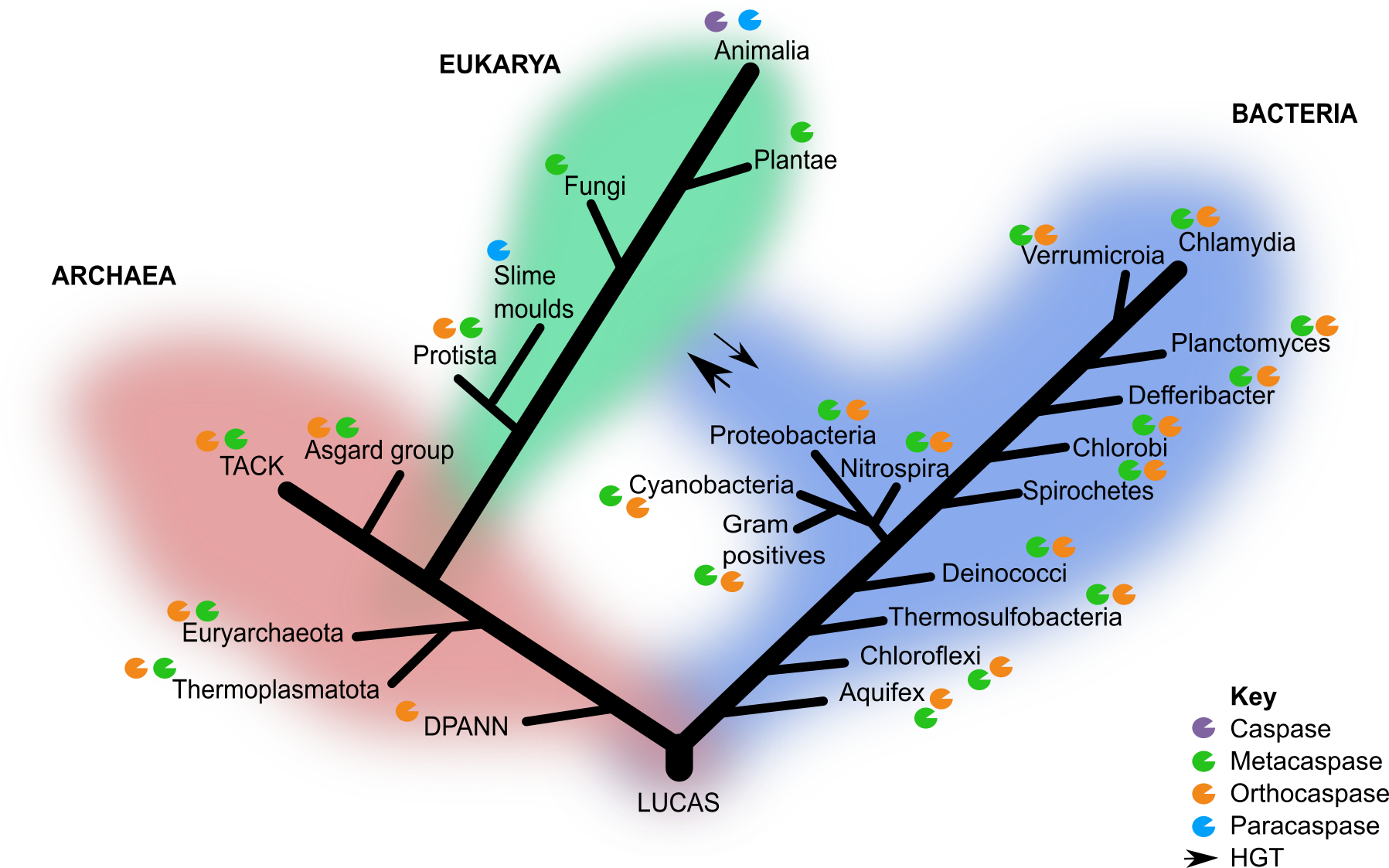
Figure 1 Canonical 3-domain tree of life with caspase homologs. Only the groups of interest included in this study are indicated. HGT events between the domain eukarya and bacteria are indicated by the arrows and thickness indicates the frequency of the event.
The conservation of OCAs in both archaea and bacteria as well as their ancient origin raise questions about their ancestral functions. Were they involved in PCD or pro-survival functions, or both? Phyletic and functional pattern analyses revealed archaeal OCAs and MCAs possess diverse C14 peptidase-accompanying domains and are likely associated with a range of cell survival (cell differentiation, cell cycle regulation, and cell metabolism) and PCD functions, as well as interactions with the environment and neighboring cells (cell adhesion, cell projection, and cell surface receptors).
An interesting finding was the distribution of C14 peptidase-accompanying domains according to the different phyla of archaea. The deep-branching phyla, Euryarchaeota and Thermoplasmatota, possessed OCAs that contained abundant C14 peptidase-accompanying domains associated with pro-survival. This also links these ancestral OCAs with cell survival activities prior to eukaryogenesis (Figure 2). OCAs and type I MCAs in the late-branching superphylum TACK possessed predominantly death domains and other C14-accompanying domains, possibly acquired via HGT (Figure 2). From this distribution, I hypothesized that archaeal OCAs were initially involved in pro-survival activities and subsequently co-opted for death-related functions in the earliest cellular organisms. This is in line with previous studies, where it was suggested that PCD evolved from effectors involved in physiological cellular processes (Klemenčič and Funk 2018).
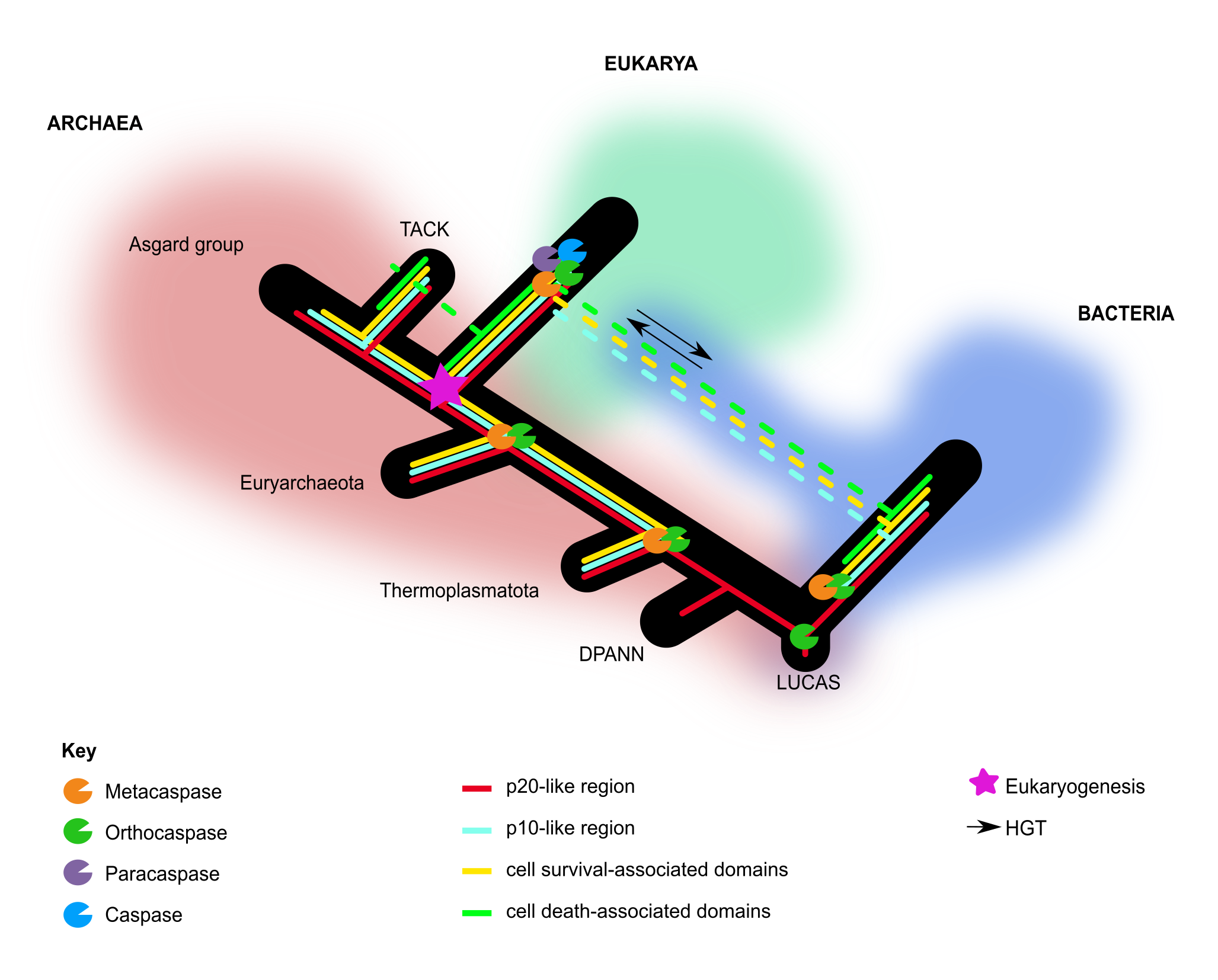
Figure 2 Proposed acquisitions of the different C14 peptidase-accompanying domains and their associated pro-survival or pro-death domains in archaea.
Current hypotheses on PCD are based on philosophical and conceptual works, including Ameisen’s ‘addiction’ and ‘original sin’ hypotheses and Durand’s ‘co-evolution’ hypothesis (Ameisen 2002; Durand 2021). The ‘original sin' postulates that PCD genetic toolkits were present since the origin of cellular life (Ameisen 2002). These data provided strong support for the ‘original sin’ hypothesis of PCD origin. CA homologs may have initially been associated with non-death functions, such as the cell cycle or adaptations to environmental stress, and subsequent refinement allowed the emergence of death-related functions. In accordance with this hypothesis, the evolutionary history of archaeal CA homologs revealed that earlier CA homologs served survival functions before being co-opted into death-related functions.
This was the first in silico study to investigate the evolutionary history of CA homologs in archaea and their putative ancestral functions. Empirical works will surely follow.
References
- Allocati, N. et al. (2015). Die for the community: An overview of programmed cell death in bacteria. Cell Death and Disease 6:1–10.
- Ameisen, J.C. (2002). On the origin, evolution, and nature of programmed cell death: a timeline of four billion years. Cell death and differentiation 9:367–93.
- Berman-Frank, I. et al. (2004). The demise of the marine cyanobacterium, Trichodesmium spp., via an autocatalyzed cell death pathway. Limnology and Oceanography 49:997–1005.
- Bidle, K.A. et al. (2010). Tantalizing evidence for caspase-like protein expression and activity in the cellular stress response of Archaea. Environmental Microbiology 12:1161–1172.
- Blackstone, N.W. and Green, D.R. (1999). The evolution of a mechanism of cell suicide. BioEssays 21:84–88.
- Choi, C.J. and Berges, J.A. (2013). New types of metacaspases in phytoplankton reveal diverse origins of cell death proteases. Cell Death and Disease 4:e490.
- Durand, P.M. (2021). The Evolutionary Origins of Life and Death. Chicago: The University of Chicago Press.
- Durand, P.M., Barreto Filho, M.M. and Michod, R.E. (2019). Cell Death in Evolutionary Transitions in Individuality. Yale Journal of Biology and Medicine 92:1–12.
- Engelberg-Kulka, H. et al. (2006). Bacterial programmed cell death and multicellular behavior in bacteria. PLoS Genetics 2:1518–1526.
- Iranzo, J. et al. (2014). Virus-host arms race at the joint origin of multicellularity and programmed cell death. Cell cycle (Georgetown, Tex.) 13:3083–3088.
- Klemenčič, M. and Funk, C. (2018). Structural and functional diversity of caspase homologues in non-metazoan organisms. Protoplasma [Online] 255:387–397. Available at: https://pubmed.ncbi.nlm.nih.gov/28744694.
- Klim, J. et al. (2018). Ancestral State Reconstruction of the Apoptosis Machinery in the Common Ancestor of Eukaryotes. G3 (Bethesda) 8:2121–2134.
- Koonin, E. and Aravind, L. (2002). Origin and evolution of eukaryotic apoptosis: the bacterial connection. Cell death and differentiation 9:394–404.
- Koonin, E. V (2009). On the origin of cells and viruses: primordial virus world scenario. Annals of the New York Academy of Sciences 1178:47–64.
- Minina, E.A. et al. (2020). Classification and Nomenclature of Metacaspases and Paracaspases: No More Confusion with Caspases. Molecular Cell [Online] 77:927–929. Available at: https://linkinghub.elsevier.com/retrieve/pii/S1097276519309505.
- Peeters, S.H. and de Jonge, M.I. (2018). For the greater good: Programmed cell death in bacterial communities. Microbiological research 207:161–169.
- Rawlings, N.D. et al. (2014). MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Research [Online] 42:D503–D509. Available at: https://doi.org/10.1093/nar/gkt953.



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in