LIQUID ORGANELLES ORCHESTRATING ASSEMBLY OF VIRUS GENOME
Published in Microbiology

Influenza A virus (IAV) contains a segmented genome composed of 8 different RNAs in the form of viral ribonucleoproteins (vRNPs). Virions do not contain more than 8 segments and each segment is included once. This agrees with seminal work showing that the assembly of IAV genome is a complex selective process1-6. Segmentation is, however, advantageous to the virus as, in combination with the high mutation rate of the viral RNA polymerase, accelerates viral evolution which is the underlying cause of yearly epidemics. In addition, it permits genetic mixing of different parental strains through re-assortment, which originates pandemic genomes associated with severe outbreaks. This has motivated the Cell Biology of Viral Infection group, at the Instituto Gulbenkian de Ciência in Portugal, for the past 6 years, to explore the host mechanisms involved in the assembly and re-assortment of IAV genomes.
The most accepted model for influenza A genome assembly predicts that vRNPs exit the nucleus to bind Rab11 vesicles (facing the cytosol) for transport to the plasma membrane7-9. During the process, random vesicular collision events sequentially assemble the genome10,11, originating viral inclusions where the 8 vRNP-types co-locate.
The team headed by Maria Joao Amorim and composed of two post-doctoral researchers (Alenquer M and Vale-Costa S), a PhD student (Etibor TA), a Master student (Sousa AL) and a laboratory manager (Ferreira F) decided to inspect this dispersed collision model. We reasoned that if intersegment interactions originated viral inclusions, cells expressing a single vRNP-type would fail to assemble such structures. To our surprise, this was not the case. Such observation led to a paradigm shift and to the proposal of a compartmentalized genome assembly model, in which infection organizes specialized compartments dedicated to genomic complex formation.

Viral inclusions thus emerge as relevant sites for human health which prompted us to investigate in detail their: (i) physical properties, (ii) inter- and intra-communication, (iii) response to environmental changes, (iv) topographic location, and (v) function. We found that these structures are membraneless organelles with liquid properties that exchange material dynamically and adapt fast to physiological changes. Interestingly, we also showed that generation of viral inclusions is spatially regulated, occurring juxtaposed Endoplasmic Reticulum Exit Sites, which suggests a cross-talk between membrane-bound and membraneless liquid organelles. This topography agrees with a recent manuscript suggesting that as vRNPs reach the cytosol, they attach (facing the cytosol) to a modified ER, from where they are captured by Rab11 for delivery to the cell surface12. However, how and where the genomic complex would assemble was not proposed.
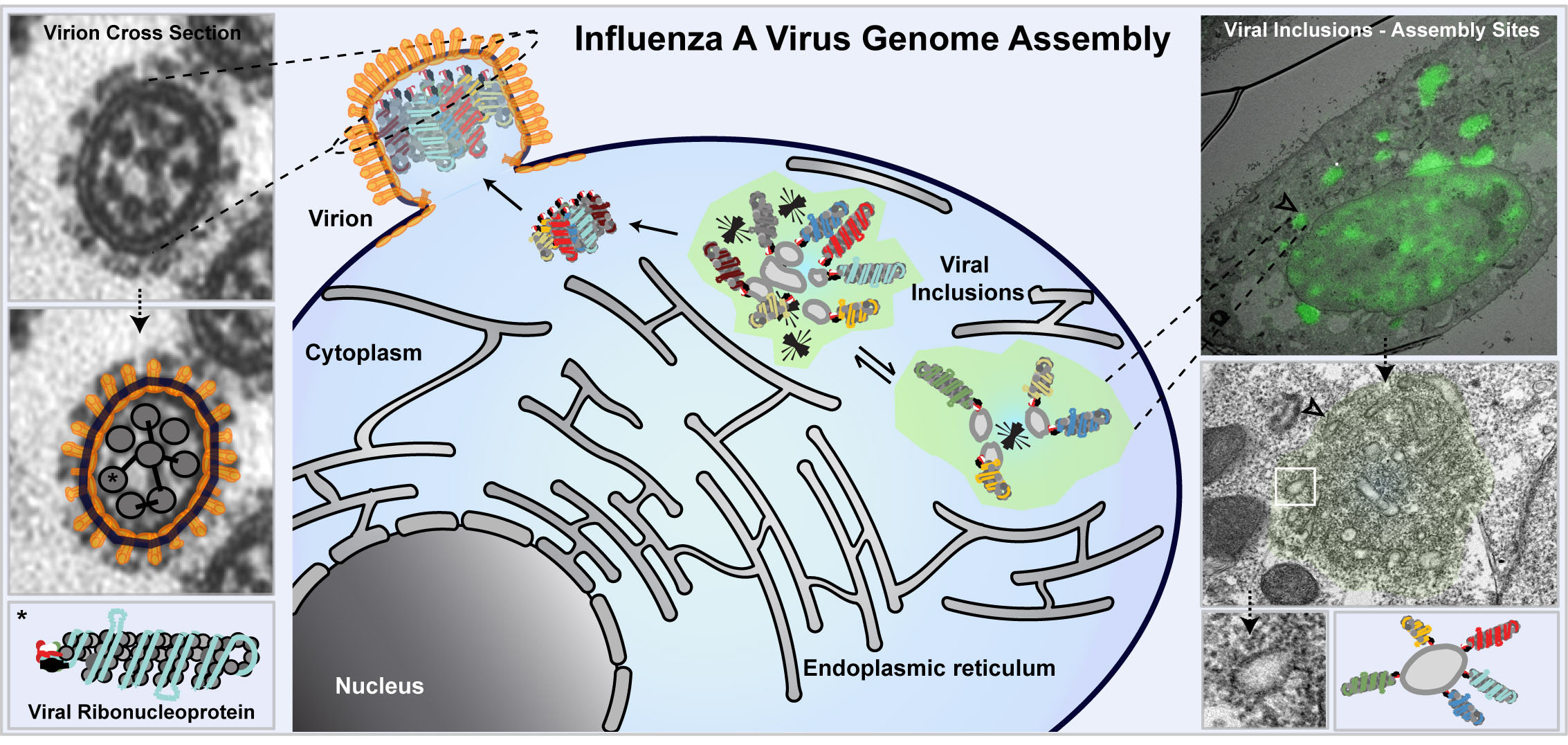
Our work shows that viral inclusions co-house vRNPs upon infection with one and also with two IA human strains. Together, the data suggest that formation of inclusions is a conserved and robust mechanism, dependent on the host protein Rab11 binding to vRNPs13, where liquid and dynamic platforms of compartmentalized vRNPs orchestrate genome assembly of epidemic and pandemic IA genomes.
REFERENCES
1. Davis, A. R., Hiti, A. L. & Nayak, D. P. Influenza defective interfering viral RNA is formed by internal deletion of genomic RNA. Proc Natl Acad Sci U S A 77, 215-219 (1980).
2. Duhaut, S. D. & Dimmock, N. J. Heterologous protection of mice from a lethal human H1N1 influenza A virus infection by H3N8 equine defective interfering virus: comparison of defective RNA sequences isolated from the DI inoculum and mouse lung. Virology 248, 241-253, doi:10.1006/viro.1998.9267 (1998).
3. Duhaut, S. D. & McCauley, J. W. Defective RNAs inhibit the assembly of influenza virus genome segments in a segment-specific manner. Virology 216, 326-337, doi:10.1006/viro.1996.0068 (1996).
4. Fournier, E. et al. A supramolecular assembly formed by influenza A virus genomic RNA segments. Nucleic Acids Res 40, 2197-2209, doi:10.1093/nar/gkr985 (2012).
5. Noda, T. et al. Importance of the 1+7 configuration of ribonucleoprotein complexes for influenza A virus genome packaging. Nat Commun 9, 54, doi:10.1038/s41467-017-02517-w (2018).
6. Amorim, M. J. A Comprehensive Review on the Interaction Between the Host GTPase Rab11 and Influenza A Virus. Frontiers in Cell and Developmental Biology 6, doi:10.3389/fcell.2018.00176 (2019).
7. Amorim, M. J. et al. A Rab11- and microtubule-dependent mechanism for cytoplasmic transport of influenza A virus viral RNA. J Virol 85, 4143-4156, doi:10.1128/JVI.02606-10 (2011).
8. Eisfeld, A. J., Kawakami, E., Watanabe, T., Neumann, G. & Kawaoka, Y. RAB11A is essential for transport of the influenza virus genome to the plasma membrane. J Virol 85, 6117-6126, doi:JVI.00378-11 [pii] 10.1128/JVI.00378-11 (2011).
9. Momose, F. et al. Apical Transport of Influenza A Virus Ribonucleoprotein Requires Rab11-positive Recycling Endosome. PLoS One 6, e21123, doi:PONE-D-11-05025 [pii] 10.1371/journal.pone.0021123 (2011).
10. Chou, Y. Y. et al. Colocalization of different influenza viral RNA segments in the cytoplasm before viral budding as shown by single-molecule sensitivity FISH analysis. PLoS Pathog 9, e1003358, doi:10.1371/journal.ppat.1003358 (2013).
11. Lakdawala, S. S. et al. Influenza a virus assembly intermediates fuse in the cytoplasm. PLoS Pathog 10, e1003971, doi:10.1371/journal.ppat.1003971 (2014).
12. de Castro Martin, I. F. et al. Influenza virus genome reaches the plasma membrane via a modified endoplasmic reticulum and Rab11-dependent vesicles. Nat Commun 8, 1396, doi:10.1038/s41467-017-01557-6 (2017).
13. Vale-Costa, S. et al. Influenza A virus ribonucleoproteins modulate host recycling by competing with Rab11 effectors. J Cell Sci 129, 1697-1710, doi:10.1242/jcs.188409 (2016).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in