Metagenomic imine reductases: enzyme discovery, screening and synthetic application
Published in Chemistry
The quest for new biocatalysts is an ongoing process, fuelled by an ever expanding availability of sequence data. Platforms such as metagenomics, which allows access to vast amounts of sequence data without the need for pre-culturing the microorganism, offers an opportunity for new biocatalyst discovery. Biocatalysts for the asymmetric synthesis of amines has been of great interest over the last twenty years, with enzyme families such as imine reductases (IREDs) and a sub-class of reductive aminases (RedAms) garnering particular attention over the last decade. We, the group of Prof. Nicholas J. Turner and other research groups have been extensively investigating this particular enzyme class, although until recently were really only investigating single or several enzymes, for example the reductive aminase from Aspergillus oryzae (AspRedAm)1.
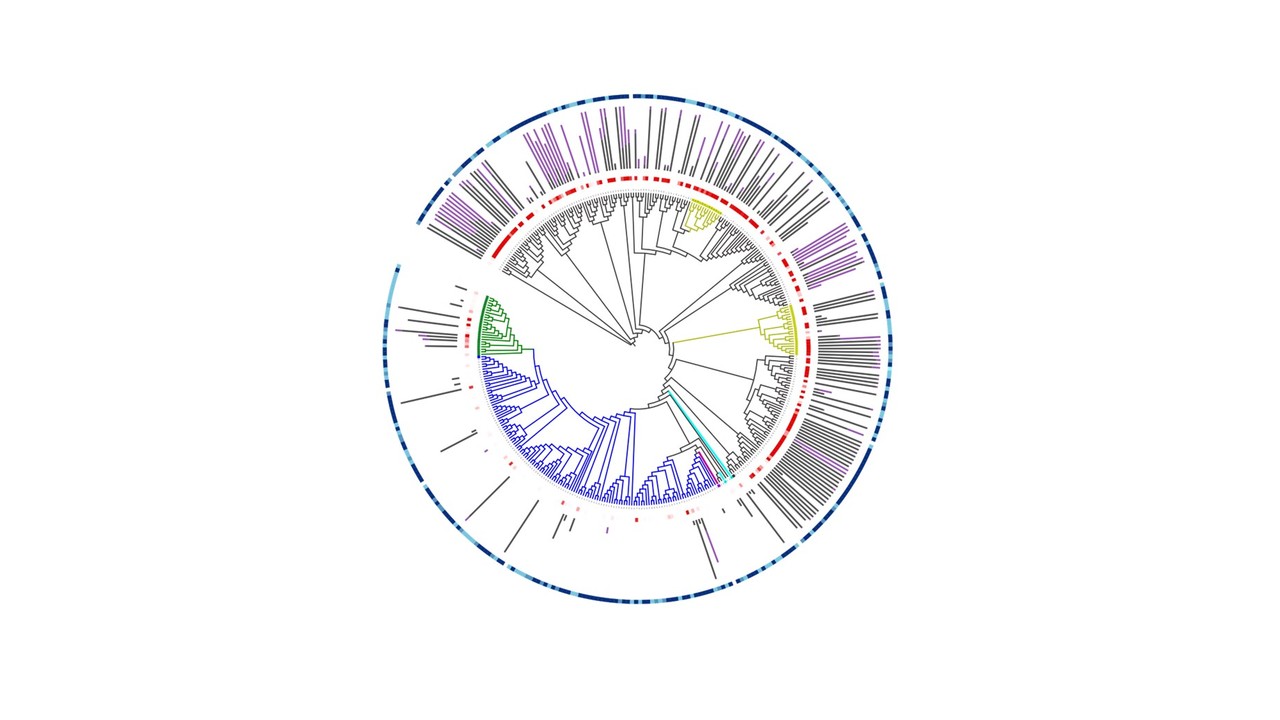
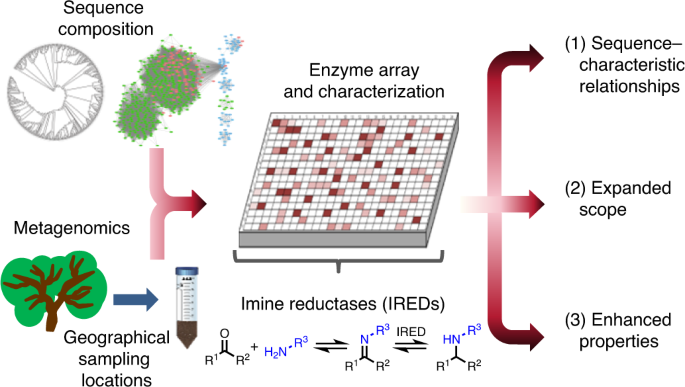
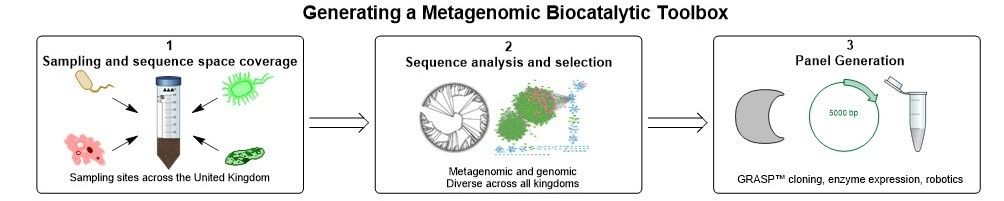
Realising that metagenomes are likely to offer a wealth of data to identify new IREDs, we collaborated in 2017 with an enzyme discovery company, Prozomix Ltd, based in Northumberland, England. The aim was to curate the largest and most sequentially diverse panel of IREDs available to date, thereby providing a biocatalytic platform for effective reductive amination reactions (Figure 1). Prozomix had sampled various geographical locations across the United Kingdom, ranging from forest to coastal areas, generating huge metagenomics collections. Query sequences such as AspRedAm and several other sequences were used to probe the metagenomes for new IREDs, with over 300 putative genes then being identified. Owing to cheap and accessible gene synthesis, these enzymes were rapidly cloned and subsequently expressed in E. coli. With further reduction in gene synthesis and sequencing costs, along with technologies to rapidly assemble metagenomes, the time frames to fabricate enzymes panels will inevitably decrease accompanied with an increase in enzyme numbers. Additionally, the 200 sequences given in the SI were chosen as the enzymes are centrally related to the findings of the article. Further sequences were chosen evenly from the phylogenetic space investigated. The sequences given allow for further exploration of new sequence space around that we have explored.
Figure 1- Generating a metagenomic biocatalytic toolbox. Several of the stages required to generate a metagenomic toolbox including the sampling of geographical locations (box 1). Sequence analysis to ensure correct dataset is obtained, including generation of sequence similarity networks (box 2). Enzyme panel generation with cloning, expression in E. coli and microtitre plate generation (box 3).
In our mind, the assembly of such a large number of proteins needed easy access to an end user. Therefore, we chose to array the lyophilised enzymes into 384-well microtitre plates (Figure 2). This gave us the opportunity to readily carry out 384-well biotransformations to be then analysed by GC or HPLC, or another analytical method. We also chose, to aid ourselves and researchers in the wider community, to develop a tetrazolium based colorimetric screen performed in the oxidative direction. The screen was useful in identifying IREDs towards our chosen synthetic applications of the enzymes. Hits identified via the screen require validation in the reductive direction. The screen can also be adapted to different conditions such as identifying enzymes operating at higher temperatures, or other reaction conditions.
Figure 2- Plate lyophilisation process. A stack of 384-well microtitre plates being freeze-dried at Prozomix Ltd.
Initially we screened the IREDs for reductive amination, targeting previously challenging areas for both carbonyl (acetophenone substrates) and amine (dimethylamine substrates) partners, generating chiral amines of industrial relevance. We then sought to expand the reductive amination scope of these enzymes into new chemical space by challenging them with β-keto esters, where products were obtained in excellent yields and purities. The use of metagenomics IREDs allowed us access to both enantiomers of the chiral amine products, without recourse to further protein engineering.
The diverse biocatalytic platform we have developed here has really changed the way we work with IREDs and other enzyme families within the Turner-Flitsch group at Manchester. The availability of these enzymes has allowed to us to identify new IREDS which catalyses previously unreported transformations. We also know that in public repositories and metagenomic collections there are still many IRED sequences yet to be explored. Who knows what additional transformations these oxidoreductases may catalyse? By combining metagenomics with retrosynthesis tools such as RetroBioCat2 and more streamlined directed evolution approaches, we can begin to access even more compounds of industrial applicability and relevance.
Find out more about metagenomic IREDs in Nature Chemistry.
References:
-
Aleku, G. A. et al. A reductive aminase from Aspergillus oryzae. Nat. Chem. 9, 961–969 (2017).
-
Finnigan, W., Hepworth, L. J., Flitsch, S. L. & Turner, N. J. RetroBioCat as a computer-aided synthesis planning tool for biocatalytic reactions and cascades. Nat. Catal. (2021) doi:10.1038/s41929-020-00556-z.
Follow the Topic
-
Nature Chemistry

A monthly journal dedicated to publishing high-quality papers that describe the most significant and cutting-edge research in all areas of chemistry, reflecting the traditional core subjects of analytical, inorganic, organic and physical chemistry.




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in