Modeling the evolution of the first multicellular life cycles
Published in Ecology & Evolution
I became fascinated with the fundamental question of multicellular development – how does a single cell develop into a multicellular organism – when I first saw it posed in one of my undergraduate courses, and it remained a common thread throughout my training as a biologist. This project introduced me to a completely different way of approaching the problem of development: instead of high-tech molecular tools, we used a simple theoretical model, and instead of looking at present-day development, we went back all the way to the origin of multicellularity – long before well-defined multicellular organisms with tightly regulated development evolved. We set out to explore what multicellularity would look like at the very beginning, and how it might be shaped by the unicellular ancestor and its ecology.
I was lucky that when I started working on this project in March 2017, my co-authors (Corina Tarnita at Princeton University and Jordi van Gestel at the University of Zürich) had been thinking about these questions for quite some time and had recently recorded their thoughts in a perspective piece in PNAS. Their perspective proposes a bottom-up approach to major transitions in evolution and in particular the evolution of multicellularity. In this approach, the ancestor takes center stage, and one asks what evolutionary innovations might emerge starting from a particular ancestor. In this spirit, my first steps were to play around with different ways to model the unicellular ancestor, before even thinking about multicellularity.

At this time, I was a master’s student at Utrecht University. My master’s program encourages students to do an internship abroad, so in September 2016 I had sent Corina Tarnita an e-mail asking whether she would be interested in working on a theoretical project together. She said yes, and we agreed that after working from Utrecht for a few months, I would visit her lab at Princeton’s EEB department to work on the project together. Working in an ecology and evolution department turned out to be a bit of a culture shock: being used to the controlled lab experiments of molecular developmental biology, it took me a while to get accustomed to being around scientists studying biology in the chaos of the real world. But three months sufficed to convert me: I enjoyed thinking about evolutionary questions instead of figuring out molecular mechanistic details, and, after I completed my master’s, I applied to Princeton’s EEB graduate program. Corina is now my PhD advisor and I study the developmental questions I’ve always been interested in from an evolutionary perspective.
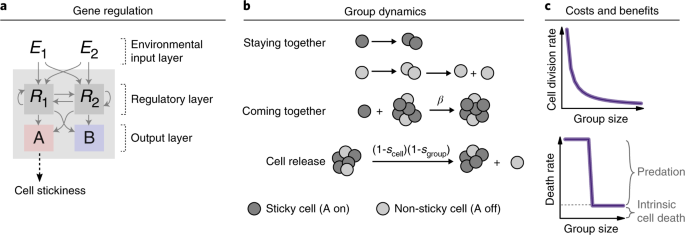
When this project first brought me to Princeton in September 2017, I was starting to get skeptical that the ancestor I had been modeling would enable us to learn about multicellularity – after all, it was not doing anything more than regulating its gene expression in response to the environment. But my co-authors thought that this ancestor would be complex enough for some interesting multicellular behaviors to emerge, yet simple enough for us to still understand what happened and why. When we added multicellularity to the model, we realized their intuition had been right. Inspired by recent work on “primitive” multicellular organisms that highlights the importance of co-option of ancestral mechanisms, we assumed that one of the ancestral genes would start allowing cells to stick to each other, in addition to its ancestral function. For example, a gene that encodes a cell surface protein that allows ancestral cells to bind extracellular entities, could, under novel environmental conditions, enable the formation of multicellular groups.
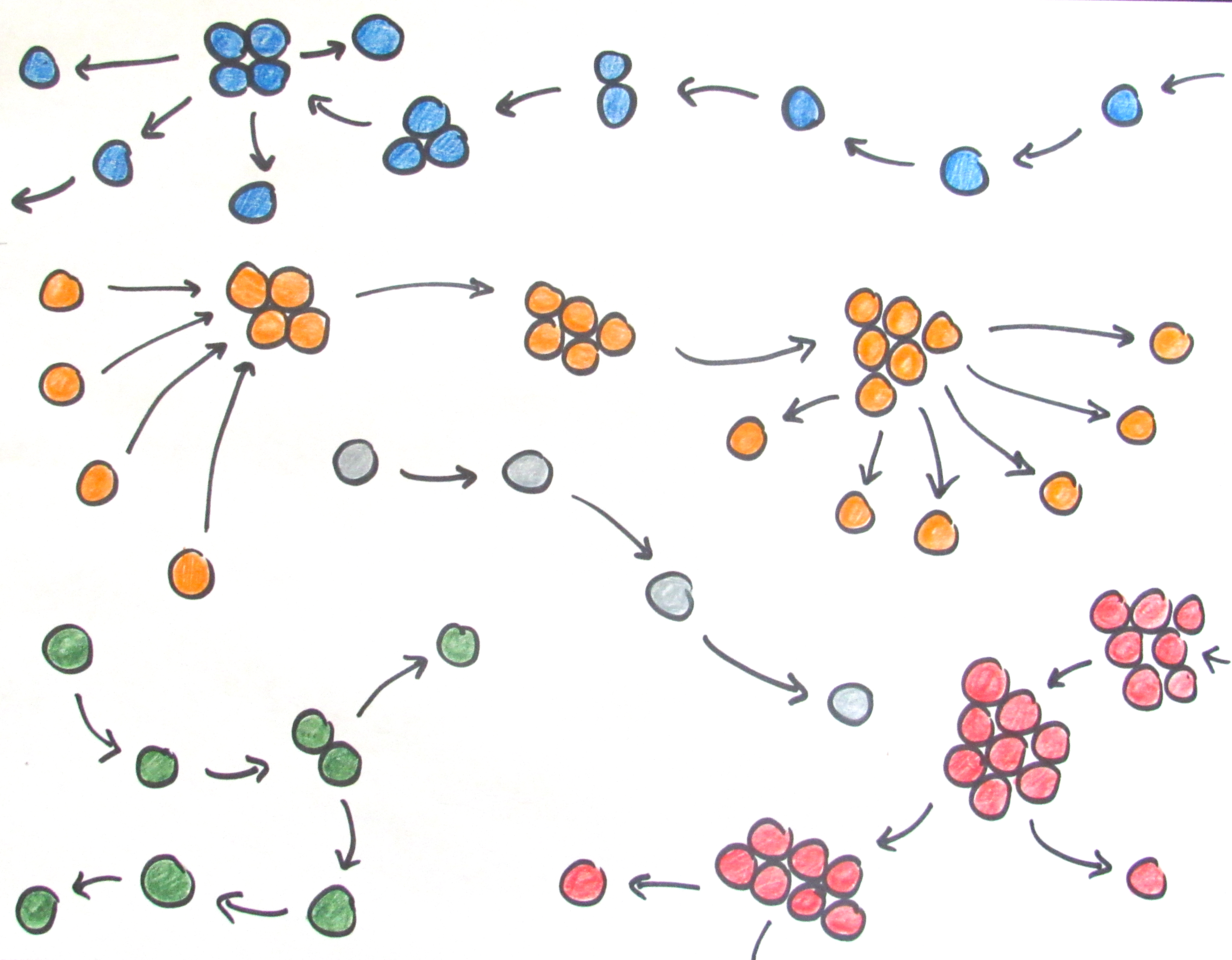
We assumed that forming groups would be costly, but that sufficiently large groups would be able to avoid predation and hence have a survival advantage. When we introduced multicellularity in our simulations, we initially saw occasional group formation: groups formed whenever the ancestor expressed its stickiness gene in response to the environment. As our simulations progressed, groups started forming continuously as cells evolved that expressed the stickiness gene all the time, allowing them to fully reap the benefits of becoming multicellular. Thus, our model could indeed account for the evolution of (obligate) multicellularity.
In some simulations, however, something unexpected happened. We found average group size to keep fluctuating over time, contrary to my expectation that group size should eventually reach an equilibrium determined by a balance between group growth (through cell divisions) and group shrinkage (through cells detaching from their group). It was only after I tracked the expression of the multicellularity gene timestep-by-timestep that I found out what was going on. At regular intervals, cells briefly switched off the stickiness gene, disrupting cell adhesion and causing complete disintegration of groups into single cells. This is what prevented the group dynamics from ever stabilizing!

While the weird fluctuations were now explained, I was confused about why groups would disintegrate willingly into single cells, and worried that this would somehow be a quirk of the way I implemented the model. But, in fact, disintegration into single cells turned out to be a clever adaptation: it allows groups to stay small (group formation costs increase with group size in the model) and propagate efficiently, without being dependent on the accidental release of cells for group reproduction. A life cycle had emerged in which groups grow for a fixed amount of time, and then release all their cells as unicellular propagules that can initiate new groups (there are ample parallels with real-world life cycles, from cellular slime molds to cyanobacteria or ichthyosporeans). This was exciting! We realized we could use our model to explore the emergence of different types of multicellular life cycles. Indeed, our model shows that, even from simple beginnings, diverse life cycles may emerge from the interplay of ancestral constraints and ecological conditions.
That our model outputs these unexpected multicellular life cycles is, for me, one of the highlights of this work. It demonstrates the value of using simple mathematical models to study big evolutionary questions, even when we do not have the empirical data to fill in the details of such a model. I could not put it any better than biologist John Tyler Bonner: “So much of [modeling] involves trying to marry empirical data with the best-fitting equations, with a great emphasis on testing the predictions of the model. [...] But here, the models are used entirely for fantasyland; it is a way of asking how it might have been in the beginning. It is our greatest tool to ask that question, free of details and frills.”
Follow the Topic
-
Nature Ecology & Evolution

This journal is interested in the full spectrum of ecological and evolutionary biology, encompassing approaches at the molecular, organismal, population, community and ecosystem levels, as well as relevant parts of the social sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Biodiversity and ecosystem functioning of global peatlands
Publishing Model: Hybrid
Deadline: Jul 27, 2026
Understanding species redistributions under global climate change
Publishing Model: Hybrid
Deadline: Jun 30, 2026

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in