Molecular profile of a killer – how COVID-19 autopsies help to understand the deadly new coronavirus disease
Published in Microbiology

March 2020: The world started to face the fact that COVID-19 had turned into a deadly pandemic. At that time, rational treatments were not available. I am a pathologist in Switzerland and used to work quietly behind the scenes and assist clinical colleagues in integrating our results into patient-management strategies. Pathologists are the doctors who, together with highly trained medical laboratory professionals, identify, validate and implement biomarker protocols and analyses to accurately diagnose patients. Suddenly, our institute received daily calls for help about any insight we could provide from autopsies of COVID-19 patients, as the scarcity of published COVID-19 autopsy cases left treating physicians at a loss about the causes of death1. We undertook the challenge to change this situation, enabled by an autopsy facility for highly infectious cases and together with colleagues across Switzerland and from Italy2. Our current lung autopsy study now allows us to trace the molecular course of COVID-19 in lungs through an integrated analysis of 34 autoptic lung tissues from 16 patients that add significant novelty to the current knowledge of the field.
While much of Europe was in lockdown, we rallied an interdisciplinary team of scientists with strong backgrounds in surgical and molecular pathology, infectious diseases, immunology and bioinformatics. Our molecular analyses of COVID-19 lung autopsy tissues resulted in a specific gene signature, with interferon stimulated genes (ISGs) as the most striking finding. ISGs are well-known from rheumatological disorders, and we found a robust up-regulation of these genes in COVID-19 lungs – but surprisingly only in about half of our deceased patients.
Along with these clear differences in ISG expression, we discovered two clear and very distinct disease patterns in autopsy lungs from fatal COVID-19. One pattern was characterized by high local expression of ISGs and cytokines, high viral load, sparse immune infiltrates, and relatively limited hemorrhagic lung damage. The other pattern showed low local expression of ISGs, low viral load, yet abundant infiltrating activated cytotoxic T cells and macrophages, massive pulmonary damage, expression of tissue regeneration markers and high local complement activation. Of note, patients with an ISGhigh profile in the lung died, on average, after a significantly shorter hospitalization period than patients with an ISGlow profile (Figure 1A). Therefore our observations allow to delineate the course of COVID-19 in the lung (Figure 1B) and are in line with a large epidemiologic study describing two distinct groups of peak lethality, one after short, and one after extended hospitalization3.
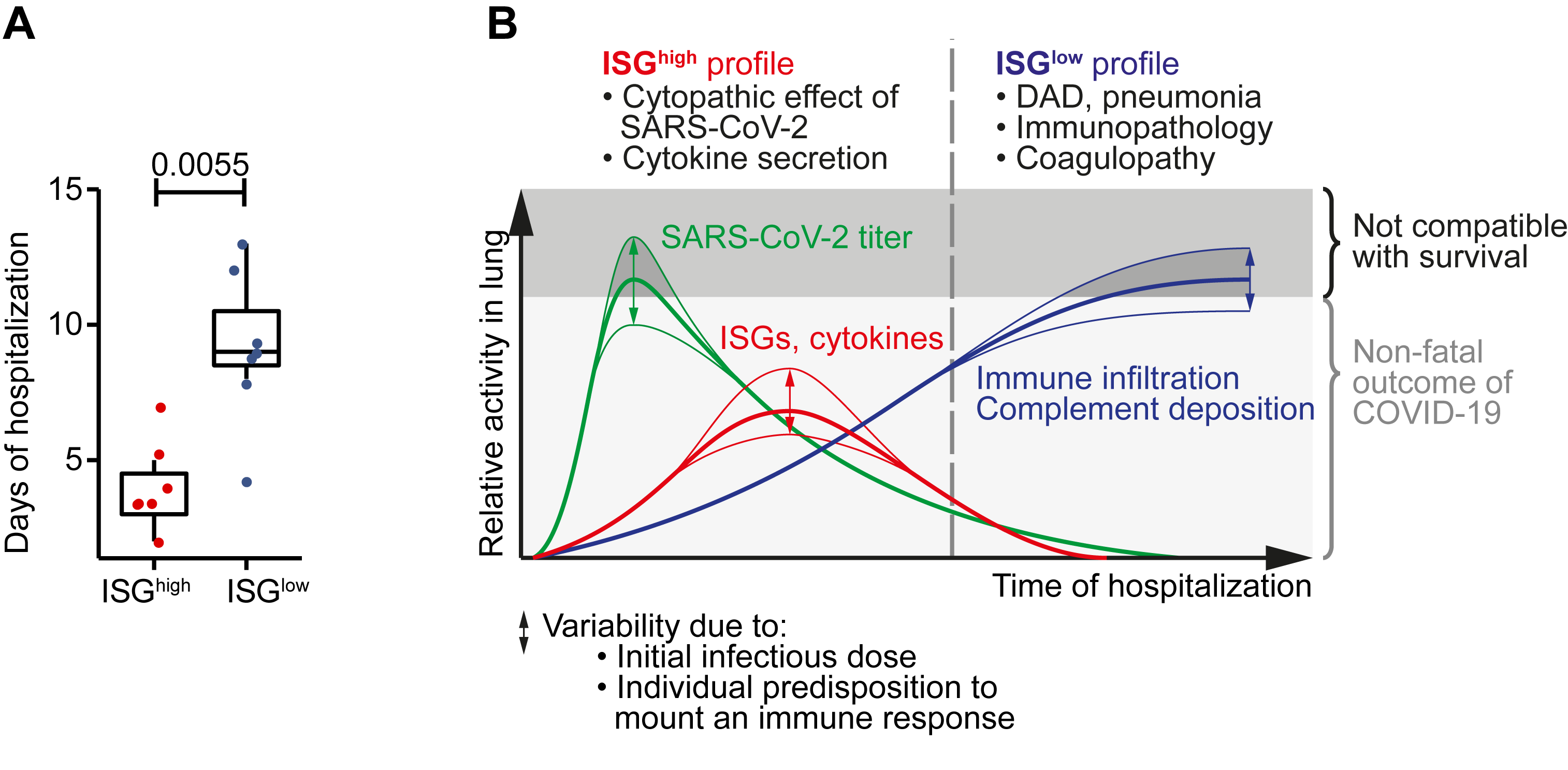 Figure 1: Main findings from an integrated analysis of COVID-19 lung autopsies.
Figure 1: Main findings from an integrated analysis of COVID-19 lung autopsies.
A Hospitalization time in ISGhigh patients versus ISGlow patients. B Schematic time course of COVID-19 lung disease based on autopsy findings. Early in the disease, an ISGhigh lung profile is observed, with high expression of cytokines and ISGs, high viral load and sparse immune infiltrates. Late in the disease, an ISGlow lung profile is observed, with low local expression of cytokines and ISGs, low viral load and strong infiltration of macrophages and lymphocytes. Patients who die early are not able to adequately control SARS-CoV-2, while patients who die late suffer from DAD and immunopathology. Infectious dose and individual predisposition to mount immune responses likely define whether or not a patient survives COVID-19.
Our findings have several implications. First, our study sharpens the clinician’s eye for two patterns of COVID-19 lung disease, and suggests two distinct molecular and cellular disease stages. Second, our results point to specific nodes of therapeutic intervention connected to these stages: Some early critical patients (ISGhigh) may benefit more from antiviral medication or antagonists of innate cytokines, while others later in the disease (ISGlow) may benefit more from therapies targeting complement activation. Third, the correlation of low viral counts and an activated CD8+ T cell signature in lungs implies that establishing CD8+ immunity should be considered in vaccination efforts. On a practical note, we used a commercially available gene expression assay for transcriptomic profiling that was originally developed for immuno-oncology (OIRRA). The accessibility of such assays could be an encouragement for hospitals around the world to conduct similar molecular profiling studies of diagnostic tissue samples from COVID-19 patients. Consequently our report aligns with the aim of the scientific community to disseminate translational research that helps improve therapy, patient outcomes and vaccination efforts.
There is a lot we still do not know about COVID-19. Still too many of our patients loose their battle against the disease. We as pathologists will continue to take that challenge. We are committed to playing our full part in the collective effort of the medical and scientific community as the COVID-19 pandemic continues to unfold. Every autopsy performed expands our opportunity to learn more and save lives. Let’s take on this challenge together!
- Ledford, H. Autopsy slowdown hinders quest to determine how coronavirus kills. Nature (2020). doi: 10.1038/d41586-020-01355-z. PMID: 32382121.
- Menter, T., et al. Post‐mortem examination of COVID19 patients reveals diffuse alveolar damage with severe capillary congestion and variegated findings of lungs and other organs suggesting vascular dysfunction. Histopathology 77, 198-209 (2020).
- Salje, H., et al. Estimating the burden of SARS-CoV-2 in France. Science 369, 208-211 (2020).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in